Introduction
In the lab "The Cell" of The Biology Lab Primer, you will:
- review the major organelles of eukaryotes
- investigate plant cell structures
- discover how chloroplasts while undergoing photosynthesis
- identify easily identifiable organelles within human cells
- conduct a gram stain to visualize plaque bacteria
Exercise A: Eukaryotic organelles
In this exercise, you will review the main structures and organelles of the eukaryotic cell. You will follow the initiation, synthesis and modification of a protein within various organelles. Organelles that are not involved directly in protein synthesis are also discussed. Below is a simplified version of eukaryotic organelles.
Organelles involved in the synthesis of a protein.
In eukaryotic cells, DNA is composed of linear strands, known as chromatids, housed in the nucleus of the cell. DNA is a genetic storage molecule that synthesizes RNA molecules: messenger RNA (mRNA), transfer RNA (tRNA), and ribosomal RNA (rRNA). The process by which DNA codes for mRNA is known as transcription. In this process, a precise segment of DNA (known as a gene) opens up and synthesizes a specific strand of mRNA. After the mRNA is synthesized, post-transcriptional modification occurs in which segments of the mRNA are removed (known as introns) and the remaining mRNA segments (exons) are reattached. This mature mRNA (composed of reattached exons) leaves the nucleus through the nuclear pore and travels into the rough Endoplasmic Reticulum (Rough ER). In the rough ER, the mature mRNA attaches to a ribosome. At the ribosome, the process of translation begins in which the mRNA codes for the synthesis of a protein.
Once the protein is completely synthesized at the ribosome, the protein leaves the rough ER through a transport vesicle. Many proteins in eukaryotic cells require further processing, which happens in the Golgi apparatus. A transport vesicle with the unmodified protein travels to the cis-face of the Golgi apparatus along the cell’s cytoskeleton network. Once the transport vesicle enters into the Golgi apparatus, the peptide bonds are broken and rearranged, creating an altered protein. In addition, identification tags are put on the protein that allow it to be placed exactly where it is designed to be. Many proteins end up on the cellular membrane and are involved in transportation of materials across the membrane. The modified protein leaves the Golgi apparatus through the trans-face and connects to the cytoskeleton network, travelling to its identified location.
That protein will continue to function until it deteriorates. Once proteins lose their functionality, the cell has an internal detection system that signals lysosomes to engulf and dissolve the protein back into its fundamental monomers, amino acids.
Organelles not directly involved in protein synthesis
While most of the organelles in a eukaryotic cell are somehow involved in the production of proteins, there are other organelles that serve other functions. Perhaps the most important of these is the mitochondrion. This organelle is involved in cellular respiration and is capable of producing up to a net of 38 ATP (the universal currency of energy of all cells) from a single glucose molecule. Prokaryotic cells that do not have any mitochondria can only produce a net of 2 ATP from a glucose molecule. For this reason, the mitochondrion has been coined “the powerhouse of the cell”.
Smooth ER doesn’t have ribosomes and thus appears smooth under a microscope. Smooth ER’s primary purpose is the production of lipids. While both the smooth ER and rough ER produce phospholipids, smooth ER also produces hormones.
Vacuoles are simply a phospholipid bilayer inside of the cell. The function of these organelles is primarily to store materials. Plant cell vacuoles are typically very large because their main function is to store water between rains. Animal cells also have vacuoles but they tend to be very small in comparison to plant cells.
In the cell diagram within the The Biology Lab Primer:
- Label the following events of protein synthesis and recycling
- Transcription
- mRNA leaves through nuclear pore
- Translation at ribosome in rough ER
- Transport vesicle shuttles unmodified protein
- Protein is modified in Golgi Apparatus
- Lysosome digests worn out protein
- Label the mitochondrion
- Label the vacuole
- Label the smooth ER
Experiment 1: Investigating plant cells
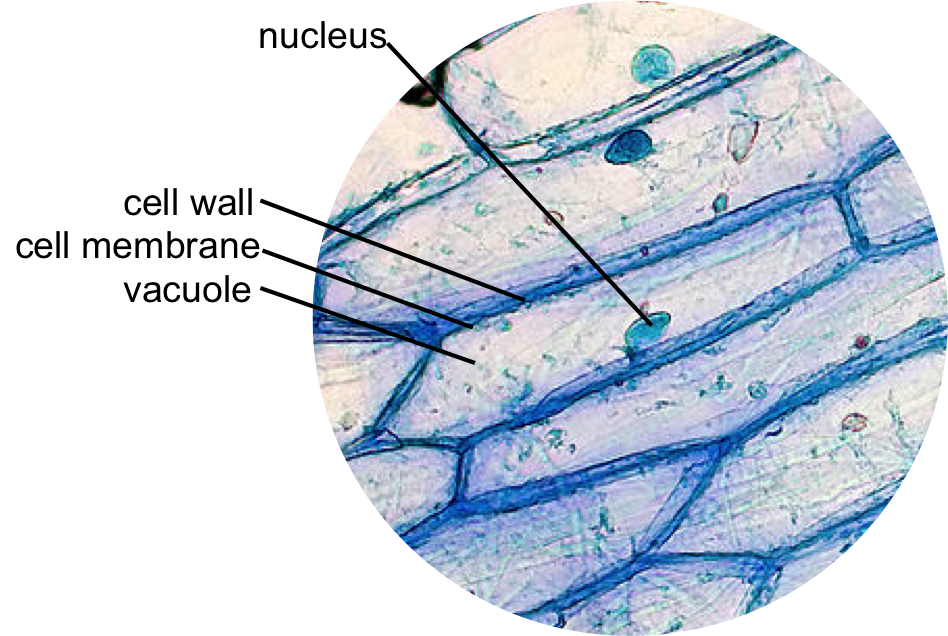
Figure 14. Epidermal onion cells under a microscope. Plant cells appear polygonal from the presence of a cell wall made of cellulose (and sometimes lignin), located just exterior of the cell membrane. The cell membrane and vacuole are typically difficult to view with this staining procedure. The vacuole is a large organelle that occupies most of the interior of the cell. An iodine stain allows the nucleus to be easily seen, typically as a orange-red structure in the middle of the cell. It is bound by a nuclear membrane. You may be able to detect the darker nucleolus within the nucleus.
Part 1: The Onion Cell
Biologists frequently study the onion cell (Figure 14) because onions are readily available and their cells provide a clear view of all the basic characteristics of plant cell structure. The onion's large cells can be seen easily under a microscope and also used to teach the fundamentals of cell biology. The skin (or epidermis) between the dormant leaves of an onion are a single cell thick, and serve as a classic representation of the internal structure of plant cells. In fact, the term "cell" came from a pioneer of microscopic biology, Robert Hooke, while looking at epidermal onion cells under a microscope. He thought these structures resembled the cells that monks would sleep in.
The onion's cell walls, like those of other plants, are rigid. Cellulose in the cell walls forms clearly defined polygonal structures.
Water within the cell walls gives the walls strength and helps plants resist the force of gravity. The cell's cytoplasm and vacuole contribute to the onion's solidity and its characteristic crisp texture.
Below the cell wall is a layer of liquid called the cytosol, primarily composed of water, salts and organic molecules. The cytosol also contains organelles, organic structures that manage all the elements of cell metabolism. The cytosol also carries inclusions, which are starches, proteins and other elements that act as building blocks for a number of functions. The nucleus, also found in this cytoplasmic layer, contains the plant's basic genetic material.
The onion has a large vacuole, which contains water and ions and produces the onion's distinctive aroma and taste.
It is likely (though not impossible) that you will not find chloroplasts in an onion cell. Onions are underground storage organs, which consist of modified leaves emerging from a very short stem. This morphology allows vegetative organs of onions to persist underground through cold winters. In spring, the modified leaves undergo rapid cell division and green leaves emerge to the surface. It may be difficult to detect chloroplasts within your onion cells. However, if you look closely you will see small, emerald-colored organelles within some cells. These are the dormant chloroplasts, which in spring rapidly replicate producing the green color of the emerging leaves.
Protocol - for use in class labs; online students refer to Figure 14
- Prepare onion wet mount.
- Quarter an onion.
- Isolate a single onion leaf.
- Break leaf in half and pull thin layer of epidermis, being careful to not let it fold on itself. If it does fold, attempt to unfold it. You may want to use forceps.
- Place the flattened onion epidermis on a microscope slide.
- Place a small drop of iodine on the epidermis. Iodine will stain the nucleus of the cell so it is visible.
- Place cover slip on top of the specimen.
- With a paper towel, remove any excess iodine.
- Place wet mount on microscope.
- With the scanning objective (4x), focus the specimen and locate a section where the epidermis is a single cell thick. This is usually found on the edges of the specimen.
- Switch to low power objective (10x). Focus in on a section of the epidermis where it is a single cell thick.
- Switch to high power objective (40x) and focus, only with the fine focus knob.
In The Biology Lab Primer, you will
- Draw a representative onion epidermal cell identifying the following structures: cell wall, cell membrane, nucleus, and nuclear membrane.
- Identify whether your onion cells have chloroplasts or not, and discuss why?
Part 2: Cytoplasmic streaming in Elodea
Elodea is an freshwater, aquatic plant native to the Americas commonly used in aquariums. With leaves only two cells thick. Elodea is a good model to study living plant cells in action. In this experiment, you will see chloroplasts moving in the Elodea cells as they begin to photosynthesize. This movement is known as cytoplasmic streaming, which distributes nutrients more evenly throughout the cell. Imagine is most of the chloroplasts within a plant cell were concentrated within a part of the cell. Molecules essential for photosynthesis (i.e. carbon dioxide) would quickly be metabolized. This would leave an unequal distribution of carbon dioxide in the cell. Movement of the organelles around the vacuole allows for a more even distribution of molecules necessary for the cell's essential biochemical pathways. It has also been suggested cytoplasmic streaming, allows greater efficiency of photosynthesis, by increasing overall light absorption.
Protocol - for use in class labs; online students refer to the video below
- Store Elodea in a dark place for at least one hour.
- Prepare a wet mount of Elodea.
- Select a leaf from an Elodea plant.
- Place the leaf on a microscope slide.
- Place a drop of water on the leaf and place a cover slip over the specimen. Push the cover slip down and use a paper towel to remove excess water.
- Focus the Elodea under scanning objective. Switch objectives to low section near the edge attempting to find a section where the cells are a single layer thick. Switch to high objective and refocus with the fine adjustment knob.
- Turn on the light source of the compound microscope to full intensity. This will allow the chloroplast to begin to undergo photosynthesis. Leave the wet mount of Elodea under the light for 15-20 minutes.
- After 15-20 minutes, reduce the light so that you may visualize the Elodea cells. You should see the chloroplasts moving in a circular pattern around the cell. This process is known as cytoplasmic streaming.
In The Biology Lab Primer, you will:
- Identify and label the major organelles of the Elodea cell. The space inside of the cell is known as the cytoplasm. In the cytoplasm, there are several green oval organelles, known as chloroplasts. In the center of the cell is a large vacuole used to store water and other molecules. Surrounding the cell is the cell wall, which appears as relatively thick boundaries between the Elodea cells. Just inside the cell wall, you may or may not be able to see the cell membrane. Nuclei are typically very difficult to during this experiment. Nuclei are typically invisible under a light microscope unless a stain is applied, which subsequently kills the cells.
- Analyze how cytoplasmic streaming occurred in your cells (or from the video for online classes). Did it happen? Why is it happening? How are the chloroplasts moving through the cell?
Experiment 2: Investigating animal cells
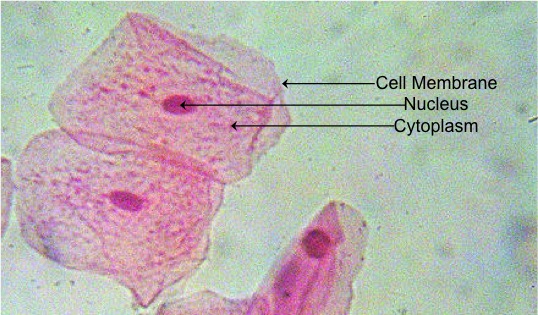
Figure 16. Stained human cheek cells. Using this very simple staining procedure, we can easily identify some of the basic structures of an animal cell. Nuclei appear as small, dark elliptical structures within the cell. If you can isolate a single cell, it will be easy to detect the boundary of the cell, the cell membrane. Between the cell membrane and nucleus is a fluid, known as the cytoplasm. Within the cytoplasm are several organelles. However, it is difficult to determine which organelles these are with this staining procedure.
Cheek cells are epithelial cells that line the interior surface of our mouths. The base layer of cells in an epithelial structure are not actually cells, but a sticky layer on which the cells anchor. The other surface of the epithelial cell touches the outside world (like skin) or an open space (like the mouth). Because of their high rate of division, epithelial cells are found tightly packed together. When you stain your cheek cells, you should be able to distinguish between the nucleus, cytoplasm, cell membrane. If you are very observant (and lucky) you may visualize the nucleolus and other organelles with in the cell.
Protocol - for use in class labs, online students refer to Fig. 16
- Using a flat toothpick, very gently scrape the inside of your cheek to obtain cheek cells.
- Spread the cells on the end of the toothpick onto the microscope slide.
- Add 1 small drop of methylene blue to the sample. Methylene blue will stain the sample, allowing visualization of the nucleus, cytoplasm, and even some organelles. Note: methylene blue will stain your hands and clothing. Wear goggles, gloves and an apron.
- Place a cover slip on the sample. Press down on the coverslip and remove excess methylene blue with a paper towel.
- Using the scanning objective (4x), focus the specimen and locate cheek cells. Change the objective to low power, refocus on a few cheek cells.
- Finally, visualize your cheek cells at high power. At this point you may need to reduce your light intensity and adjust the condenser aperture.
Experiment 3: Investigating plaque bacteria
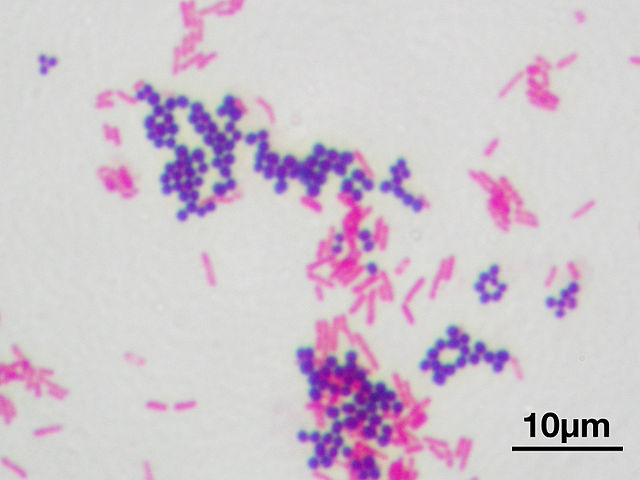
Gram staining is a method of differentiating bacterial species (gram-positive and gram-negative). Gram staining differentiates bacteria by allowing detection of peptidoglycan abundance. All bacteria have cell walls made of peptidoglycan. Gram positive bacteria simply have more peptidoglycan than gram negative bacteria. The cell types differ in other ways as well. Gram-positive bacteria have a single cell membrane, with a thick cell wall composed of peptidoglycan. This thick cell wall appears dark purple following a gram stain due to the high abundance of peptidoglycan In contrast, gram negative bacteria have a thin cell wall (also made of peptidoglycan) sandwiched between two cell membranes. The thin cell wall appears pink in gram negative bacteria following a gram stain. It was originally hypothesized that gram positive and gram negative formed two monophyletic groups of bacteria, due to their cell wall similarities within each group. However, recent phylogenetic analyses indicate gram positive and negative bacteria do not neatly fall into two separate categories. It should be noted that the gram stain is ineffective in identifying bacteria beyond an initial investigation. Many different species are found in each group. Individuals of certain species (gram variable bacteria) can actually appear to be both gram positive and gram negative. Other species (gram indeterminate) do not accept the gram stain and can not be visualized with this procedure.
Protocol - for use in class labs, online students refer to the video above and Figs. 17-18
- On a clean slide, place one small drop of deionized water.
- With a clean toothpick, scrape your teeth near the gum line.
- Mix the scraping in a drop of water on the slide.
- Spread this mixture into a thin film on the slide. Place the slide on the slide warmer for about 5 minutes until the water has completely evaporated from the slide.
- Place the slide in the Gram staining tray, and add 4 drops of crystal violet stain (Fig. 4).
- Leave the stain on the slide for 1 minute.
- Wash off the stain with a very gentle stream of water using your squirt bottle.
- Blot the stained slide with bibulous paper. Do not smear it!!!
- Place 4-8 drops of iodine solution. Let it sit for one minute.
- Gently wash the iodine off so as not to remove the bacteria.
- Blot your slide with bibulous paper.
- Place 4-8 drops of 95% alcohol on your specimen for 10 seconds. Rinse with a gentle stream of water.
- Place 4-8 drops of safranin solution to counterstain. After 30 seconds, wash off with a gentle stream of water.
- Place a cover slip on your slide.
- Place your slide on your microscope. Bring the specimen in focus with the scanning (4x), low (10x) and high (40x) objectives. Once your specimen is in focus at high power, place a drop of immersion oil on the slide and visualize your specimen under oil immersion power. You may see your own cheek cells. Bacteria cells are approximately 1/100th the size of a eukaryotic cell.
Copyright © 2016 Jason S. Walker. All rights reserved.