Chapter: The Genetic Code: Transcription and Translation (draft)
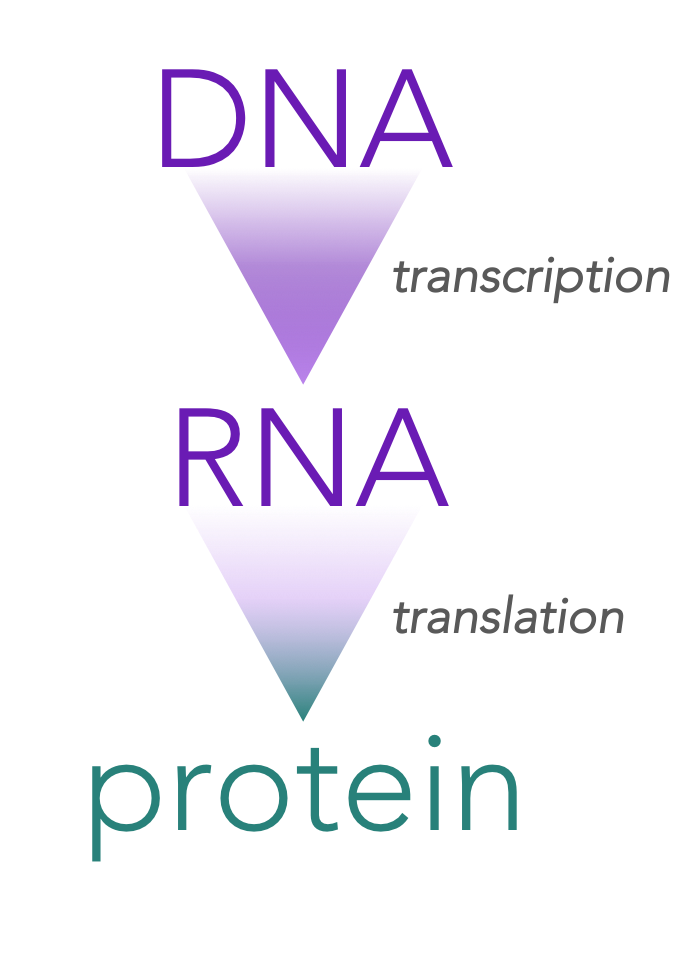
Figure 1. The Central Dogma of Molecular Biology.
The central dogma of molecular biology
James Crick (cofounder of DNA’s secondary structure) proposed that DNA is an informational storage molecule capable of replicating itself. Further, he proposed that the information that was transmitted had to be “read” by a manufacturing body within the cell which put amino acids together in a specific sequence ultimately synthesizing a protein. This became known as the central dogma of molecular biology.
The central dogma predicts that DNA serves as a template for the direct synthesis of a messenger RNA (mRNA) molecule, in a process known as transcription. Secondly, mRNA is “read” at a ribosome by transfer RNAs (tRNAs) , which work together to assemble a specific chain of amino acids, which collectively assemble to generate a protein, a process is known as translation.
Exceptions to the dogma
Figure 2. Retroviruses represent an exception to the central dogma.
Messenger RNA is just one of seven major different types of RNA. Some are also involved in protein synthesis (like transfer RNA). And DNA directly codes for these RNA molecules. So the information flow in this case would be simply DNA to RNA. The other major exception to this dogma, the information flow is reversed. Some viruses for example have genes composed of DNA. When these viruses infect a cell, the viral RNA synthesizes DNA. So in this way, the information flow would be from RNA to DNA. But even though there are exception to the dogma, the central dogma of molecular biology encompasses The most important flow of information for life. DNA codes for RNA and that RNA codes proteins.
RNA replication is the copying of one RNA to another. Many viruses replicate this way. The enzymes that copy RNA to new RNA, called RNA-dependent RNA polymerases, are also found in many eukaryotes where they are involved in RNA silencing. RNA editing, in which an RNA sequence is altered by a complex of proteins and a "guide RNA", could also be considered an RNA-to-RNA transfer.
The triplet code
Once biologists understood the dogma, they understood the general pattern of information flow in the cell. The next challenge was understand how the sequence of bases in a strand of messenger RNA code for the sequence of amino acids in a protein. What is the genetic code? What are the rules that specify the relationship between the sequence of nucleotides in DNA and the sequences of amino acids in a protein? George Gamow suggested a code based on logic. He suggested that each code word contains three bases. His reasoning was based on the observation that there are 20 amino acids.
Since there are only four unique nucleotides in DNA and 20 amino acids, a combination of base pairs was required to code for the amino acids. If an amino acid was based on a single nucleotide, then there would only be four amino acids. With the same logic, Gamow surmised that the code could not be represented by a combination of two nucleotides, because 4x4 is 16 and there are 20 amino acids. The code must be a three base code (or triplet code), because it is the simplest code that allows for the 20 known amino acids: 4 X 4 X 4 = 64. This suggests that there could be up to 64 unique amino acids. However, there are only 20 amino acids.
Properties of the code
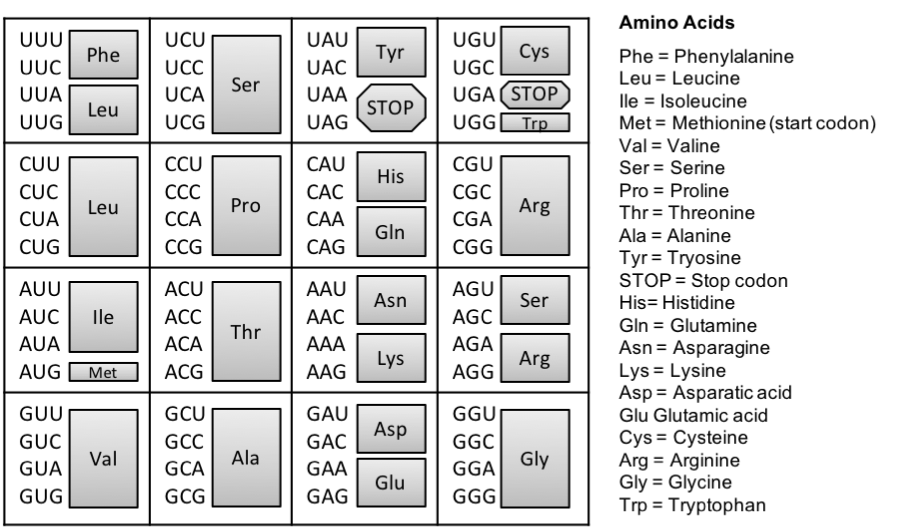
Figure 3. The genetic code. The triplet code mRNA directly codes for the assembly of amino acids that make up a protein. To identify the amino acid coded by the mRNA sequence, locate the mRNA triplet code (codon), the grey box to its right represents the corresponding amino acid. For example, CCC indicates the amino acid Proline (Pro).
There are far more possibilities of amino acids provided by a triplet code, than the number of amino acids (20) we see in nature. Therefore, it is said that the code is redundant, meaning that amino acids can be coded by more than one triplet code. For example, the triplet codes of CCU and CCC of mRNA code for the same amino acid: proline. In fact, all amino acids are coded by more than one triplet code except for methionine and tryptophan. Further investigations indicated that a specific triplet code always coded for the same amino acid. In other words, the code is unambiguous. For example, the triplet code of AUG in mRNA always codes for methionine. Amazingly, the code works exactly the same for all living organisms, from bacteria to plants and animals! While there are very few exceptions to this, the consistency of the code across widely variable organisms hints that we all stem from a single common ancestor. The code is universal. Lastly, the code is conservative. If the first two base pairs of the mRNA are the same but the third is different, there is a high likelihood (but not an absolute certainty), that the will code for the same amino acid.
The group of the three bases that species a particular amino acid is called a codon. And according to Gamow’s triplet hypothesis, each codon is made of three nucleotides. And each gene is defined by a start codon and a stop codon. The start codon has been identified, and it is the same start codon for every single gene of every single organism on Earth. In contrast, there are three stop codons.
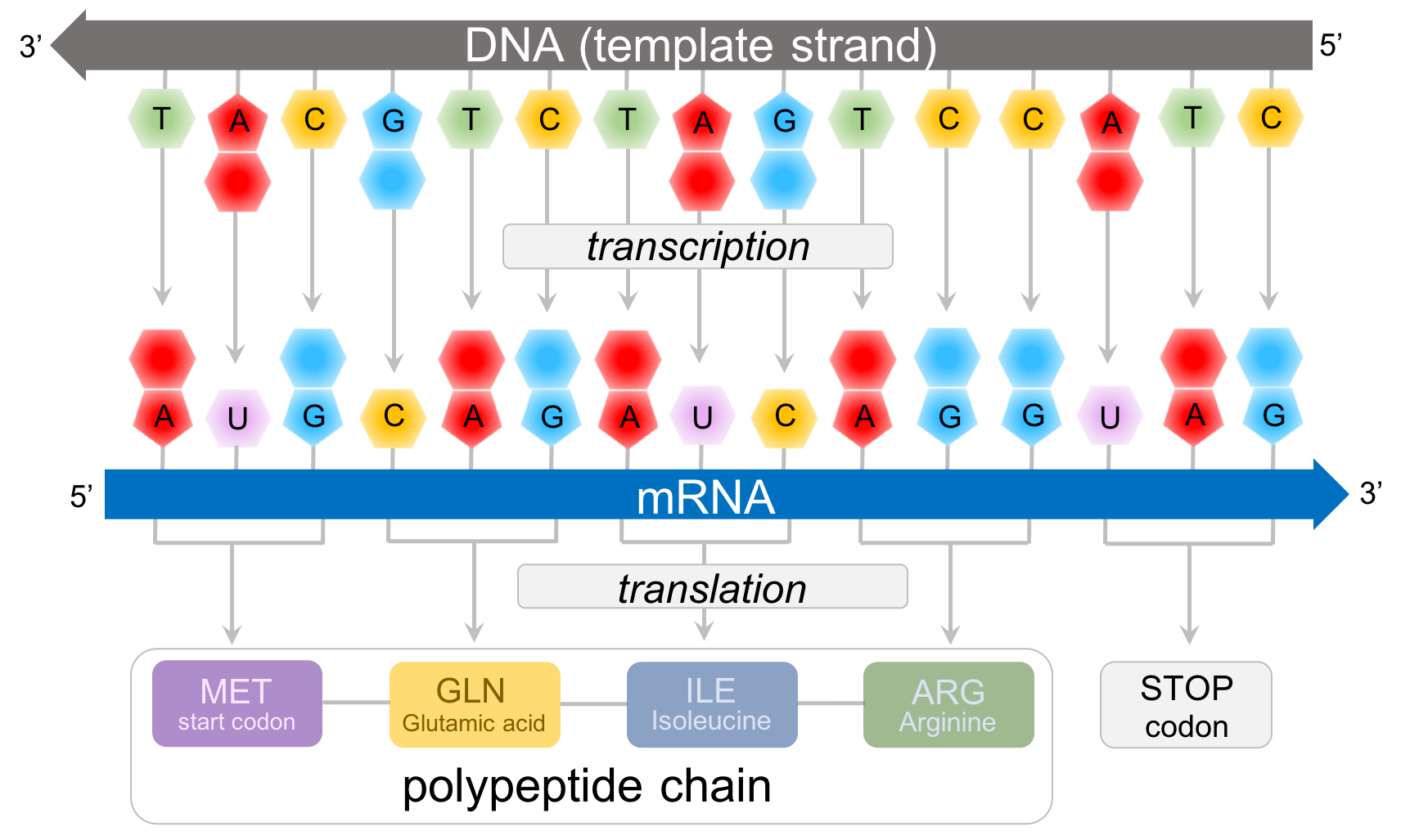
Figure 4. Predicting polypeptide chains from DNA. In this example, the template strand of DNA (the strand that transcribes into RNA) is: 3' - TAC GTC TAG TCC ATC - 5'. This is transcribed into the mRNA strand: 5' - AUG CAG AUC AGG UAG- 3'. Consulting the amino acid chart (Fig. 4), we can predict the sequence of amino acids for this protein: methionine(START CODON)-glutamic acid-isoleucine-arginine-STOP.
Predicting proteins from DNA
Once the code was deciphered, any sequence of DNA can be read in order to determine the sequence of amino acids, or a polypeptide chain (Fig. 4). In eukaryotes, DNA is composed of many chromosomes. Each chromosome is made up of multiple genes (along with other non-transcribing regions). Each gene synthesizes an mRNA, which is then transcribed into a protein. For the segment of DNA that makes up a gene, only one strand synthesizes the mRNA, known as the template stand. The other strand of DNA doesn't synthesize mRNA is called the non-template strand, or more commonly the coding strand. The beginning of a gene is defined by the three bases of the template strand, TAC, which is transcribed into the start codon, AUG. So far as we know, all living organisms have the same start codon for every protein created. The next three deoxyribonucleic acids are transcribed into the next codon. In Fig. 4, the deoxyribonucleic acids (GTC) are transcribed into the mRNA codon (CAG), which is eventually translated into the amino acid, Glutamic acid. Repeat this sequence until you reach one of the three STOP codons which, as you will see below, does not code for an amino acid. Rather it codes for a termination factor which ends the process of translation, resulting in a protein.
Mutations
Figure 5. Point mutations. Point mutations are a change to a single deoxyribonucleotide, which happens during a mismatch during DNA replication. Point mutations can affect the eventual protein by changing an amino acid in the polypeptide chain.
Mutations are permanent changes in an organism’s DNA, a modification in the cell’s information archive. Mutations are important in evolution, because they are the only know mechanism that actually creates new alleles. New alleles can create different proteins and consequently different cellular functionality, and serve as the origin of biodiversity. Mutations can either affect an organism’s fitness, an organism's ability to survive and reproduce, or not. Mutations increase fitness are termed beneficial, whereas those that decrease fitness are said to be deleterious. Mutations that have no affect on an organism’s fitness are said to be silent mutations. Most mutations are neutral or slightly deleterious. Mutations can be put into two categories. Point mutations are when a single nucleotide changes and chromosome-level mutations occur with the addition, deletion or modification of chromosome.
Point mutations
Figure 6. Genotypes determine phenotypes. A change in a single deoxyribonucleotide can change the sequence of amino acids, which can have an effect on the organism's phenotype.
Point mutations occur when DNA’s proofreading mechanism (DNA polymerase) fails to correct a mismatched base pair before the finalization of DNA replication, the process in which DNA copies itself. This results in a single base change in one of the newly synthesized DNA strands (Fig. 5). There are two resultant consequences of point mutations. Point mutations that result in change in the amino acid sequence are known as replacement mutations or missense mutations (Fig. 6). A change in the amino acid can (and often does) change the functionality of the protein it codes for, which can change the organisms fitness: either positively, negatively or not at all. Whereas, silent mutations are point changes that don’t change the amino acid sequence, because the DNA transcribes an mRNA which codes for the same amino acid as the original DNA strand. For example, if there was a change in the template strand of DNA from TAA (transcribed to the codon AUU) to TAT (transcribed as AUA), the resultant amino acid following translation would be isoleucine for both. Silent are most common when the third nucleotide in a codon is altered, highlighting the conservative property of the code.
In a species of mouse (Fig. 6), a point mutation occurred in the past at a single base pair, changing the final product of the protein resulting in a different phenotype of fur color, a missense mutation. Where the dark mouse has an arginine amino acid in its protein at a specific location, the white mouse has a cysteine. This single change in a DNA molecule is enough to cause a change in the phenotype of the mouse, and has caused members of the same species to live in different environments, a first step to becoming different species.
Chromosome-level mutations
Figure 7. Chromosome-level mutations.
Chromosome-level mutations are major changes to the DNA of eukaryotes, with the addition, deletion or movement of segments of chromosomes or even entire chromosomes. Nearly all of these mutations occur as a mistake during nuclear division (either during meiosis or mitosis) and nearly all of them negatively affect an organism's fitness. Inversion is an alteration of a single chromosome's structure, when a segment of the chromosome is detached, inverted and reattached to the same chromosomes. All genes in the inverted section are no longer serve as a template for the original proteins they transcribed for. This is due to the sequence of nucleotides are complete reverse of the original DNA, and transcription only happens in a single direction. Translocation occurs when a segment of one chromosome is removed and reattached to another chromosomes. Potentially, the translocated genes can adequately transcribe so long as no inversion has occurred.
During the cell cycle, the chromosomes replicate and are separated into different nuclei during mitosis. Following mitosis the duplicate nuclei are separated into two cells. Sometimes mistakes happen during this process. Occasionally, the duplicated chromosomes never separate, leaving an duplicate copy of chromosomes in the cell. This is known as polyploidy, and is very rare in animals. However, it is relatively common in plants and can form the origin of a new species as polyploidy plants are reproductively isolated from diploid plants. Typically during meiosis each set of replicated chromosomes is split in half, eventually producing gametes. Occasionally, an one of the replicated chromosomes doesn't properly segregate during meiosis. Once fertilized, the zygote (and eventual organism) has an additional chromosome in its cells. In humans, Down's syndrome is cause by the presence of an extra copy of chromosome 21. Two chromosome 21s came from one parent, while one came from the other. Most humans have two copies of each chromosome, one from their mother and one from their father, known as homologous chromosomes. Homologous chromosomes are similar is size and have the same sequence of genes, but can differ in the alleles they carry. Humans have 23 pairs of homologous chromosomes, 23 from the mother and 23 from the father for a total of 46 chromosomes.
Transcription
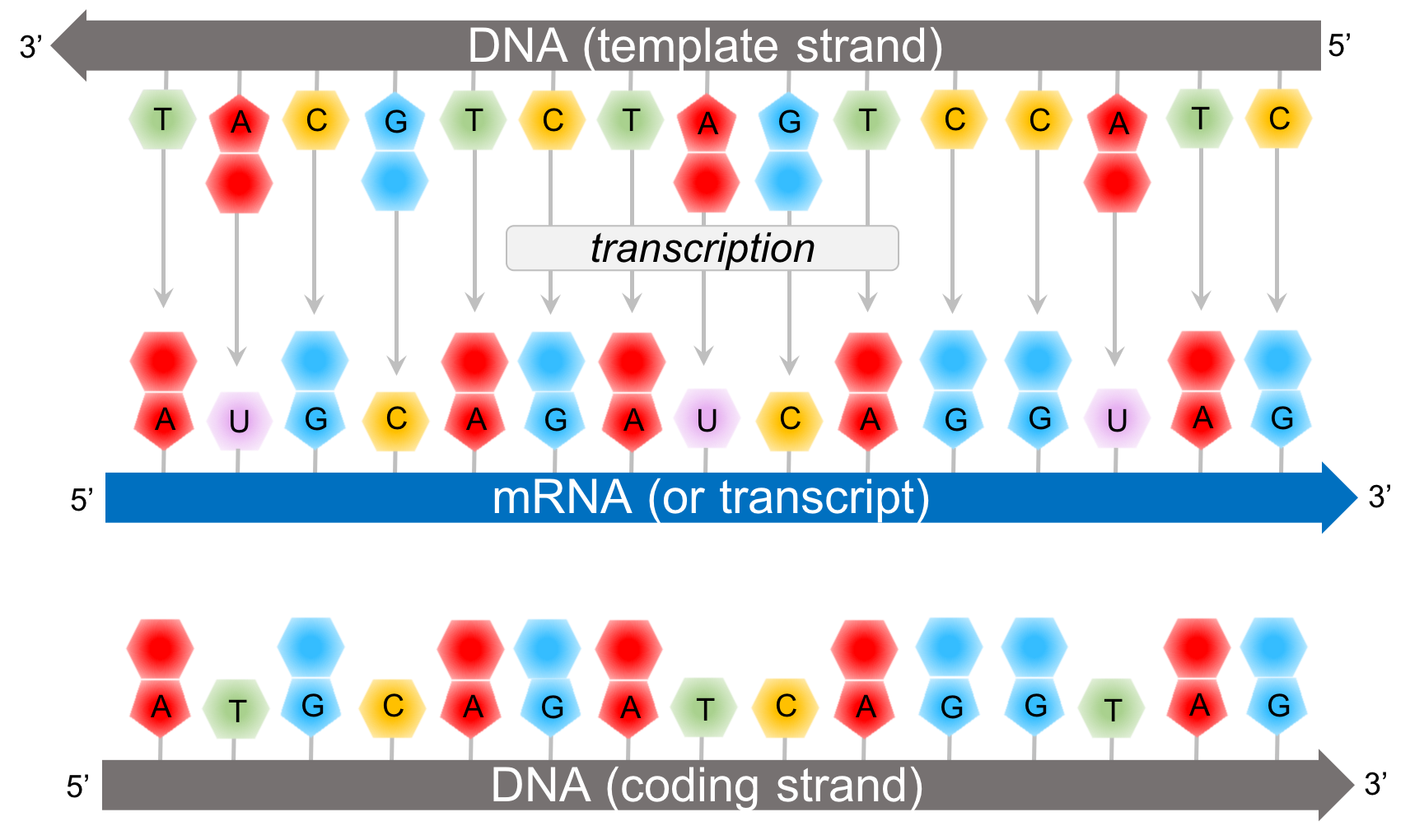
Figure 8. Transcription creates a transcript, or mRNA, according to complementary base pairing of the template strand of DNA.
In transcription, a segment of DNA (known as a gene) synthesizes mRNA. RNA are polymers composed of a chain of ribonucleotides. Ribonucleotides contain the sugar ribose; whereas deoxyribonucleotides (of DNA) contain the sugar deoxyribose. While DNA is double stranded and RNA is single-stranded, RNA contains the nitrogenous base uracil (U) where DNA would have of thymine (T). For a specific gene only one of the DNA strands, the template strand, actively synthesizes a strand mRNA, known as a transcript. The other strand of DNA, the coding strand, is not involved in transcription. However, the coding strand of DNA is more similar to the mRNA since both the coding strand and the transcript are complimentary of the template strand. However, it doesn’t match it exactly. The ribonucleotides of the transcript have the sugar ribose, and where the coding strand would have the nitrogenous base, thymine, the transcript has uracil.
During transcription, ribonucleotides bond to the template strand based on complementary base pairing via hydrogen bonds. The ribonucleotides then bond together with a phosphodiester bond just like DNA is bonded.
Initiation of Transcription
In prokaryotes, transcription is initiated by the attachment of a protein known as a sigma. The sigma attaches to one strand of the DNA (the template strand) at a very specific location. In bacteria, several sigmas exists and each one initiates the transcription of a specific sequence of DNA (or gene). Once this sigma protein attaches to the DNA molecule, it serves to guide the RNA polymerase down the template strand. The sigma protein recognizes and binds to what is deemed the promoter sequence. The promoter sequence is a specific group of base pairs. Once the sigma binds to the DNA, transcription begins. There are several different sigmas. Each one is unique and initiates the synthesis of a specific gene, or in some cases several different genes. While there are several sigmas, each for different gene complexes, RNA Polymerase is the same molecule that connects to all the different sigmas. RNA Polymerase adds ribonucleotides to the template strand based on complementary base pairing, generating an mRNA.
Figure 9. Steps of transcription in prokaryotes.
The sigma protein first opens DNA’s double helix at the promoter section of the DNA strand. Then the template strand of the DNA is threaded through the RNA polymerase. Incoming RNA nucleotides come through a channel in the sigma protein and pairs with the complementary bases of the DNA’s template strand. At this point the RNA polymerase is functional and the begins to work. And once that happens the sigma disconnects from the DNA chain. This defines the beginning of the elongation phase of transcription.
Once the appropriate sigma is attached, RNA Polymerase attaches to the sigma protein. After successful attachment, the sigma guides the DNA into place inside of the RNA Polymerase. As the DNA is thread through the RNA Polymerase, hydrogen bonds are split between the the DNA molecule, by a zipper. Once DNA is inserted in to RNA Polymerase, ribonucleotides enter an entrance portal into the RNA Polymerase and match up with the D-nucleotides based on complementary base pairing. Similar to DNA base pairing, cytosine-containing deoxyribonucleotides (D-cytosine) pair with guanine containing ribonucleotides (R-guanine), D-guanine pairs with R-cytosine, and D-thymine pairs with R-adenine. Different from DNA base pairing, D-adenine pairs with R-uracil. Through another portal in the RNA Polymerase, emerges the developing mRNA. Once a few ribonucleotides are synthesized by RNA Polymerase, the sigma protein is removed. Once the sigma is removed, it can be reused to initiate transcription.
Elongation of Transcription
Elongation in transcription is fairly straight forward. The RNA polymerase zips along the open DNA molecule matching up complementary ribonucleotide base pairs from the template strand of the open DNA (A-U, T-A, C-G, and G-C). After the sigma is removed, RNA Polymerase continues to unzip template and coding strands of the the DNA, and ribonucleotides are bonded via phosphodiester linkages based on complementary based pairing determined by the template strand of DNA. The incoming DNA enters into an intake portal and the strands are separated by an internal zipper. As the DNA passes the zipper, the hydrogen bonds reattach between the coding and template strand and the DNA double helix leaves through an exit portal. Ribonucleotides enter in through another intake portal and are combined via complementary base pairing to the template strand of DNA. The ribonucleotides are bonded to each other via phosphodiester linkages, forming an emerging . Ribonucleotides are continuously added to the 3’ end of the developing RNA strand. The 5’ end of the RNA strand leaves through another exit portal of the RNA Polymerase.
Termination of Transcription
In bacteria, once RNA Polymerase transcribes a specific sequence of ribonucleotides from the DNA template strand, transcription ends (or terminates). When this sequence is synthesized, a section of the RNA bends back on itself forms a short double helix based on complementary base pairing. This forms a RNA hairpin. This hairpin forces the RNA to separate from the DNA and the RNA Polymerase detaches and the opened DNA reattaches based on complementary base pairing
Figure 10. Steps of transcription in eukaryotes and RNA splicing.
Transcription in Eukaryotes
Fundamentally, transcription in eukaryotes is similar to transcription in prokaryotes with a few exceptions. In bacteria, RNA Polymerase can synthesize any RNA molecule. In eukaryotes, there are three different RNA Polymerases (I, II, and III). RNA Polymerase I is primarily responsible for the synthesis of ribosomal RNA (rRNA), the molecule that makes up ribosomes. Most eukaryotic RNA Polymerase are RNA Polymerase II. RNA Polymerase II is responsible for synthesizing mRNA, making it the only RNA Polymerase capable of transcribing protein-coding genes. RNA Polymerase III is responsible for synthesizing transfer RNA (tRNA). During translation, tRNAs read the messages from the mRNA and link a specific amino acid sequence generating proteins.
Where bacterial transcription is initiated by a sigma protein, RNA Polymerases in eukaryotes require a group of proteins known as basal transcription factors. Like sigma in prokaryotes, once the basal transcription factors attach to the DNA, its respective RNA Polymerase attaches and transcription begins. The elongation process is virtually identical in prokaryotes and eukaryotes. However, termination of transcription differs between prokaryotes and eukaryotes. In eukaryotes, a short sequence in the DNA signals the attachment of an enzyme downstream of active transcription. This enzyme cuts the emerging RNA, leaving the RNA Polymerase.
In eukaryotes, pre-RNA is made up of regions of mRNA that code for amino acids (known as exons) and regions of mRNA that don’t code for amino acids. Before the mRNA can be functional the introns must be removed in a process known as RNA splicing, or post-transcriptional modification.
Post-transcriptional modification of mRNA in eukaryotes
In bacteria, transcription from DNA to mRNA is a direct pathway. However in eukaryotes once mRNA is synthesized by RNA Polymerase II, the mRNA goes through further modification (Fig. 11). The product following transcription is known as a primary transcript (or pre-mRNA). Before mRNA travels outside the nucleus, the mRNA is shortened by cutting out specific sections of mRNA and reattaching the remaining sections back together. This process is known as RNA splicing and the resulting, modified mRNA is known as mature mRNA. Segments of the mRNA that are respliced back together are known as exons (because they exit the nucleus); while the segments of mRNA that are removed from the pre-mRNA are known as introns. The exons (which collectively make up the mature mRNA) leave the nucleus through a nuclear pore and travel to a ribosome in the cytosol and begin the process of translation.
RNA splicing is processed by hybrid protein-RNA complexes known as small nuclear ribonucleoproteins (or snRNPs). RNA splicing begins when a primary snRNP binds to a guanine R-nucleotide (G) adjacent to an uracil R-nucleotide (U) at the 5’ end of the pre-mRNA. This marks the exon-intron boundary. Another secondary snRNP reads from 5’ to 3’ down the mRNA and when it comes in contact with an adenine (A), and it attaches at that point. This point represents the intron-exon boundary. Once the primary and secondary snRNPs are attached other snRNPS attach to those, in a complex known as a spliceosome. Collectively the spliceosome breaks the G-U bond of the primary snRNP and the bond between the adenine (A) of the secondary snRNP and its adjacent R-nucleotide. Since U and A are complementary bases, the spliceosomes places them in close contact with each other, generating an intron loop. Nucleotides of the intron loop are disassembled into their monomers, ribonucleotides, and are recycled for future transcriptional events. Exons are spliced back together generating a mature mRNA.
Translation in prokaryotes
Figure 11. Contrast of transcription and translation in prokaryotes and eukaryotes.
Transcription is the process of creating mRNA from DNA; translation is the process of converting the genetic information of mRNA into proteins. Since prokaryotic DNA is not bounded by a nucleus, translation in prokaryotes (i.e. bacteria) occurs before transcription is complete. Translation and transcription happen simultaneously. Ribosomes are adjacent to transcribing DNA, allowing the ribosomes begin translation before transcription is terminated. This allows for translation of proteins to be more efficient in prokaryotes than eukaryotes.
Translation in eukaryotes
In eukaryotes, transcription and modification of mRNA happens exclusively in the nucleus. After mRNA processing, the mature mRNA travels out of the nucleus through a nuclear pore. In the cytosol, the body of the cell outside the nucleus, the mature mRNA attaches to a ribosome and goes through translation, eventually producing a protein. Most ribosomes are attached to the rough endoplasmic reticulum. However there are several ribosomes within the cytosol itself, as well.
This separation of transcription and translation provides a greater control over gene regulation, specifically by the removal of introns from the pre-mRNA. It is hypothesized that the removal of introns is a defense against expressing ancient retroviral genes, a key field of study in HIV research. It is also hypothesized that eukaryotic DNA is less susceptible to mutations than prokaryotes, due to the physical barrier of the nuclear envelope between the DNA and the cytosol.
Transfer RNA
In addition to mRNA, another important RNA molecule is the transfer RNA, known as tRNA, tRNA is the molecule that bridges the genetic code with the a specific protein. Each transfer molecule is attached to a specific amino acid. And each amino acid has three base pairs attached to the opposite end of it. At the ribosome, the three base pairs of the tRNA join up with the complement of the three base pairs of the mRNA. So the three complimentary base pairs of the transfer RNA are known as an anticodon; whereas the triplet code of the messenger RNA is known as a codon. Each anticodon of tRNA links with a specific amino acid is combined to it complementary codon of mRNA at the ribosome. And then the amino acids are linked together by peptide bonds into a growing peptide chain.
Ribosome
Figure 12. Steps of translation.
The ribosomal complex is made of another RNA (ribosomal RNA) and proteins. These make up two structure. The small subunit holds the mRNA in place during translation while the large subunit is where the peptide bonds form. And the large subunit has three distinct chambers: A, P and E. In general, the function of the ribosome is to synthesize proteins. First, the amino acid connected to a tRNA enters the ribosome at the A site. As another tRNA molecule comes into the A site, the other tRNA molecule moves over three base pairs and a peptide bond is formed between the two amino acids. As another tRNA molecule comes into the ribosomal complex, the other two tRNAs move 3 bases, and the oldest tRNA exits the ribosome at the E site. Just remember APE.
Initiation of Translation
Let’s take a closer look at the process translation (Fig. 12). The mRNA has a special section of it right before the start codon that the ribosome recognizes, known as the ribosome binding site. Translation begins with the start codon on the mRNA. Connected to the tRNA of the start codon is an amino acid, called f-Met. Once the f-Met tRNA binds to the small subunit of the ribosome, the large subunit of ribosome binds to the small unit, and translation is ready to begin.
Translation begins when an mRNA connects to the small subunit of a ribosome. Ribosomes are made up of proteins and another type of RNA, ribosomal RNA (or rRNA). Initiation of translation begins when rRNA binds to a specific sequence of the mRNA, known as the ribosome binding site. This connection is based on complementary base pairing of adjacent ribonucleotides of rRNA and and mRNA, which is guided into place by special proteins known as initiation factors. One of the initiation factors also serves as a docking station for the first tRNA to connect to the start codon of mRNA, which is AUG for the synthesis of all proteins. tRNAs have a complementary triplet code that connects to the codon of the mRNA, known as an anticodon (Fig. 12). The anticodon of the initial tRNA is UAC. Attached to the initial tRNA is the amino acid, methionine (met). Once this tRNA is attached to the small subunit, the large subunit of the ribosome attaches to it and elongation of translation begins.
Elongation of Translation
The next tRNA enters the A site due to complementary base pairing of the codon of the mRNA and the anticodon of the tRNA. Once the codon-anticodon pairing is successful, the new tRNA in the A site is positioned such that the amino acid it is carrying is adjacent to the amino acid already present in the P site. This proximity encourages a peptide bond to form between the two adjacent amino acids, the beginning of a polypeptide chain.
attaches to its complementary codon in the A site of the large subunit of the ribosome.
Second, a peptide bonds forms at the P site. Next the whole ribosome complex moves down three base pairs. The tRNA in the A site moves to the P site and the tRNA of the P site move to the E site, and the tRNA at the E site leaves the ribosomal complex. This process is known as translocation.
Termination of Translation
Translation is terminated by one of three stop codons. Once the ribosome encounters one of these stop codons, it causes a specific protein, known as a release factor, to enter the ribosome and it causes the release of the release of the polypeptide chain. Also at this time, the large ribosome separates from the small subunit.