Taxonomy and Classification
Exercise A: History of Taxonomy
Classification is in our nature. It can be argued that humans have been categorizing the natural world since before the advent of speech. The ability to discriminate aspects of our world is inherently necessary for our survival. In 1758, Swedish biologist Carl Linnaeus accomplished the amazing feat of classifying nearly all known organisms, published in his magnum opus Systema Naturae, categorizing all living organisms as either plants or animals (Fig 1). Systema Naturae marked the beginning of modern classification, and we still use many of the customs he developed.
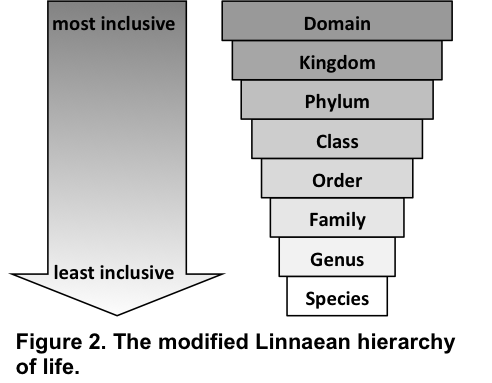
For example, Carl Linnaeus developed a hierarchical system for organizing living organisms (Fig. 2 & 3). The highest rank (most inclusive) was given to kingdom, followed subsequently by phylum, class, order, family, genus, and species. Species was Linnaeus’s least inclusive level of classification and includes all organisms of similar morphologies that can interbreed and produce viable offspring.
Given the vast scope of his work, Linnaeus completely ignored classifying single-celled organisms. Antonie van Leeuwenhoek was a pioneer of microscopy and the first person to identify single–celled organisms in 1674. He called these cells animalcules, which is Latin for “little animals.” Prior to this, the presence of single-celled organisms was completely unknown and his work was met with great skepticism. His first discoveries were of relatively large microorganisms we now call protists, unicellular organisms with a nucleus. Van Leeuwenhoek never stopped perfecting his microscope, and in 1676 he was the first person to identify and describe a bacterium, a very small microorganism lacking a nucleus and true organelles. Realizing Linnaeus’s oversight, his predecessors classified these microorganisms either as plants (if they were green) or animals. In 1866, microorganisms got a major upgrade when Ernst Haeckel proposed that unicellular organisms should be in their own kingdom: Protista. All multicellular organisms were still classified either as plants or animals (Fig. 4).
When microscope technology was enhanced during the 19th century, it became clear that microorganisms were actually two separate groups, based on their internal makeup. In 1925, Édouard Chatton argued that certain microorganisms are more closely related to plants and animals than they were to other microorganisms. He argued that organisms with the presence of nucleus should all be placed in a large group (collectively known as Superkingdom Eukaryota), whether they are unicellular or multicellular. He suggested that all other microorganisms that lack a nucleus were in a separate kingdom: Monera (Fig. 5). This theory was not well received by Chatton’s contemporaries, who argued that unicellular organisms are more closely related to each other than to any multicellular organisms (regardless of the internal makeup of the cells). Chatton’s realizations were not truly appreciated and widely accepted until the 1960s.
In 1969, Robert Whittaker recognized an additional kingdom: Fungi (Fig. 6). Prior to Whittaker’s classification, fungi were hypothesized to be in the Kingdom Plantae, due to the growth form of mushrooms coming up from the ground, similar to plants. He argued that fungi were most similar to animals because both groups are heterotrophic (they can not produce their own food); whereas plants are autotrophic (they can produce their own food). Further research has supported this hypothesis.
Also during the 1960s, Carl Woese and his colleagues began to compare RNA sequences from a wide variety of prokaryotic and eukaryotic organisms. What they found shocked the scientific community (Fig. 7). Their work suggested that certain groups of prokaryotes are actually more evolutionarily distant than the groups of eukaryotic organisms, such as plants and animals, are to each other. Their evidence suggested that all eukaryotic organisms belong in one major monophyletic group (Eukarya), while prokaryotes had to be split into two monophyletic groups (Bacteria and Archaea). They inferred that all living organisms should be placed in three domains (a newly invented level of hierarchy above Kingdom): Bacteria, Archaea, and Eukarya. These results were highly controversial, and acceptance of Woese’s findings was slow. However, they were nearly universally accepted by the 1980s.
Analysis of Woese’s phylogeny provides an interesting twist. Bacteria and Archaea are prokaryotes, which all lack a true nucleus and membrane-bound organelles; whereas all members of Eukarya have a nucleus encasing linear DNA and many membrane-bound organelles. While Bacteria and Archaea are more similar morphologically, Woese’s research suggests Archaea is genetically more closely aligned with Eukarya. This suggests that Archaea is actually more evolutionarily similar to Eukarya than it is to Bacteria, even though Archaea is more morphologically similar to Bacteria.
In the 21st century, many scientists have accepted Woese’s three-domain system as the highest order of classification of life on Earth. However, the classification of members within Eukarya has been dramatically altered based on analyses of DNA since Whittaker (Fig. 8). Protists (single-celled eukaryotic organisms) are no longer thought to be a monophyletic group. In other words, not all protists form a single, cohesive group. Evidence also suggests that multicellular organisms (plants, animals, and fungi) are a part of different eukaryotic groups that also include single-celled organisms. This indicates that multicellularity originated more than once in Eukarya.
In 2005, the International Society of Protistologists divided Eukarya into six “supergroups” based on genetic analyses (Fig. 8). Animals and fungi were included into the supergroup, Opisthokonta, alongside their suspected unicellular (and sometimes colonial) cousin the choanoflagellate. Amoebozoid protists fall into two supergroups. Traditional amoebas with lobose pseudopodia form the Amoebozoa, while amoeboids with either filose or reticulose pseudopodia form Rhizaria.
Plants were grouped with the supergroup, Archaeaplastida, which included other photosynthetic organisms, green algae and red algae, which have unicellular, colonial, and multicellular growth forms. Photosynthetic organisms are also found in two other supergroups. Excavata are flagellated protists, some of which are photosynthetic (i.e. Euglena). Diatoms are unicellular organisms that serve as the ocean’s primary source of energy via photosynthesis. Diatoms and many other unicellular photosynthetic organisms, along with multicellular brown algae, form the group Chromoalveolata. This group is widely variable, and the validity of this grouping is currently under serious scrutiny.
Exercise B: Classification of Hardware
In this exercise, you will practice the process of classifying different pieces of hardware in a process similar to how Linnaeus classified organisms of the natural world during the 18th century. Linnaeus grouped organisms based on physical similarities. Such morphological similarities are known as synapomorphies. He hypothesized that organisms which looked more similar, are more closely related. This principle of logic is known as parsimony, and is still used as the primary classification tool. After determining how different types of organisms are evolutionarily related, scientists create a phylogenetic tree (Fig. 9).
These phylogenetic trees are visualizations of ancestor-descendent relationships through time. The fewer linkages that exist between taxa on a phylogenetic tree, the more closely related they are. In the example in Figure 9, taxa B and C are more closely related than taxa A is to taxa B or taxa C.
Creating a Hardware Phylogeny
You will create a phylogenetic tree using parsimony to classify ten (10) pieces of hardware in a hierarchal fashion based on synapomorphies (similar to Fig 10).
- Acquire 10 different pieces of hardware.
- Determine how you would classify the various pieces of hardware into a phylogeny.
- On scrap paper, Include synapomorphies at each fork (technically a node).
- Synapomorphies are descriptive terms (i.e. long, spiraling, bent, 3mm long).
- Do not use nominative terms (i.e. screw, nail, bolt).
- Be sure that your branching is dichotomous (branching in twos) - never more than two and written in plain English.
- At the terminal nodes, name your hardware based on the customs of binomial nomenclature.
- The names must be in Latin. Use a translation application such as Google Translate to assist you.
- There must be two names:
- The first name corresponds to the higher order group (genus), which typically encompasses more than one type of organism (i.e. Homo). It must be capitalized.
- The second name corresponds to a specific name identifying that type of organism as a unique group (species) from other members of its genus (i.e. H. sapiens vs. H. erectus). The species name is not capitalized.
- When naming your hardware, name put the genera name first and the species name second. For example, if you were naming a short screw, you should name it in Latin as “screw short”, which translates to “Cochlia brevis”.
- There must be two names:
- The names must be in Latin. Use a translation application such as Google Translate to assist you.