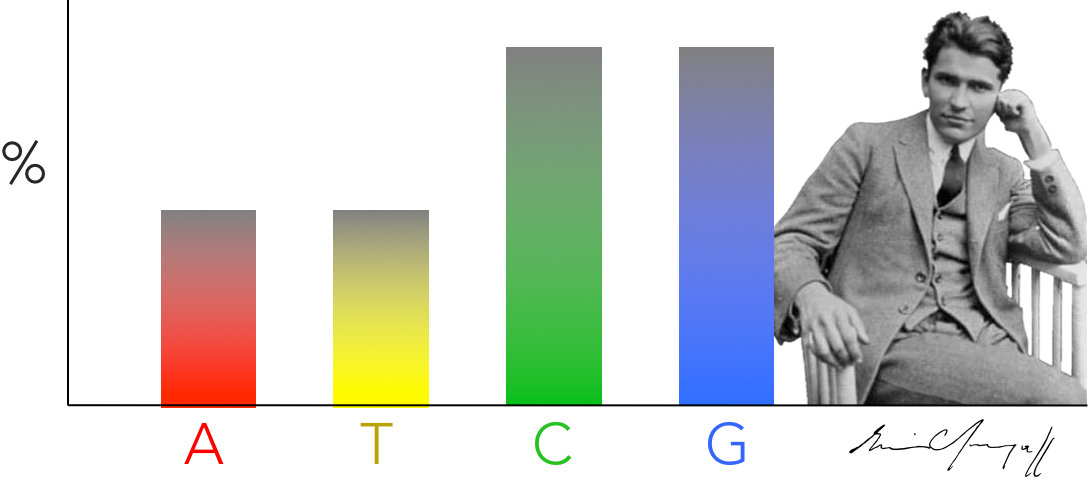Chapter 3: The Central Dogma of Molecular Biology
Lecture Video: The Central Dogma of Molecular Biology
Lecture Slides: The Central Dogma of Molecular Biology
Study Guide: The Central Dogma of Molecular Biology
Animation: The Central Dogma of Molecular Biology
Lab: Nucleotides, Nucleic Acids and Proteins
The Central Dogma of Molecular Biology
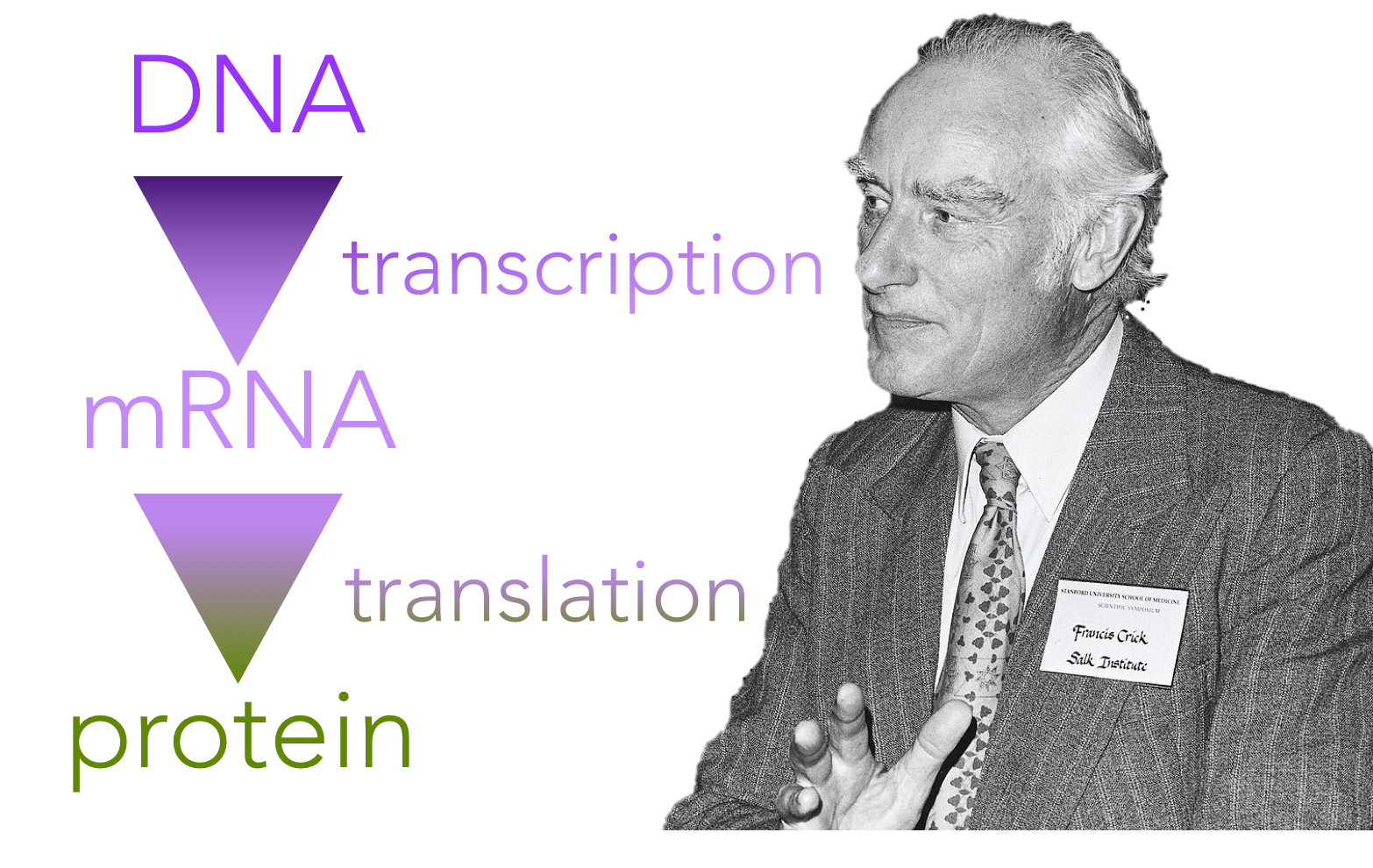
Figure 1. The central dogma of molecular biology. Coined by Francis Crick, the central dogma of biology states that DNA codes for the production of proteins, though indirectly through an intermediary molecule, RNA.
As our understanding of biological molecules increased in the 20th century, researchers discovered that all living organisms share a genetic code. In 1956, Francis Crick proposed that DNA is an informational storage molecule capable of replicating itself. Further, he proposed that the information that was transmitted had to be “read” by a manufacturing body within the cell which puts amino acids together in a specific sequence ultimately synthesizing a protein. This became known as the central dogma of molecular biology.
Specifically, DNA serves as a template for the direct synthesis of a messenger RNA (mRNA) molecule, in a process known as transcription (Fig. 1). Secondly, mRNA is “read” at a ribosome by transfer RNAs (tRNAs), which work together to assemble a specific chain of amino acids, which collectively assemble to generate a protein, in a process known as translation. Proteins are the cell’s internal machinery. Similar to parts of a car, each protein has a specific three-dimensional shape that determines its function. Any change in the shape potentially changes the function of the protein.
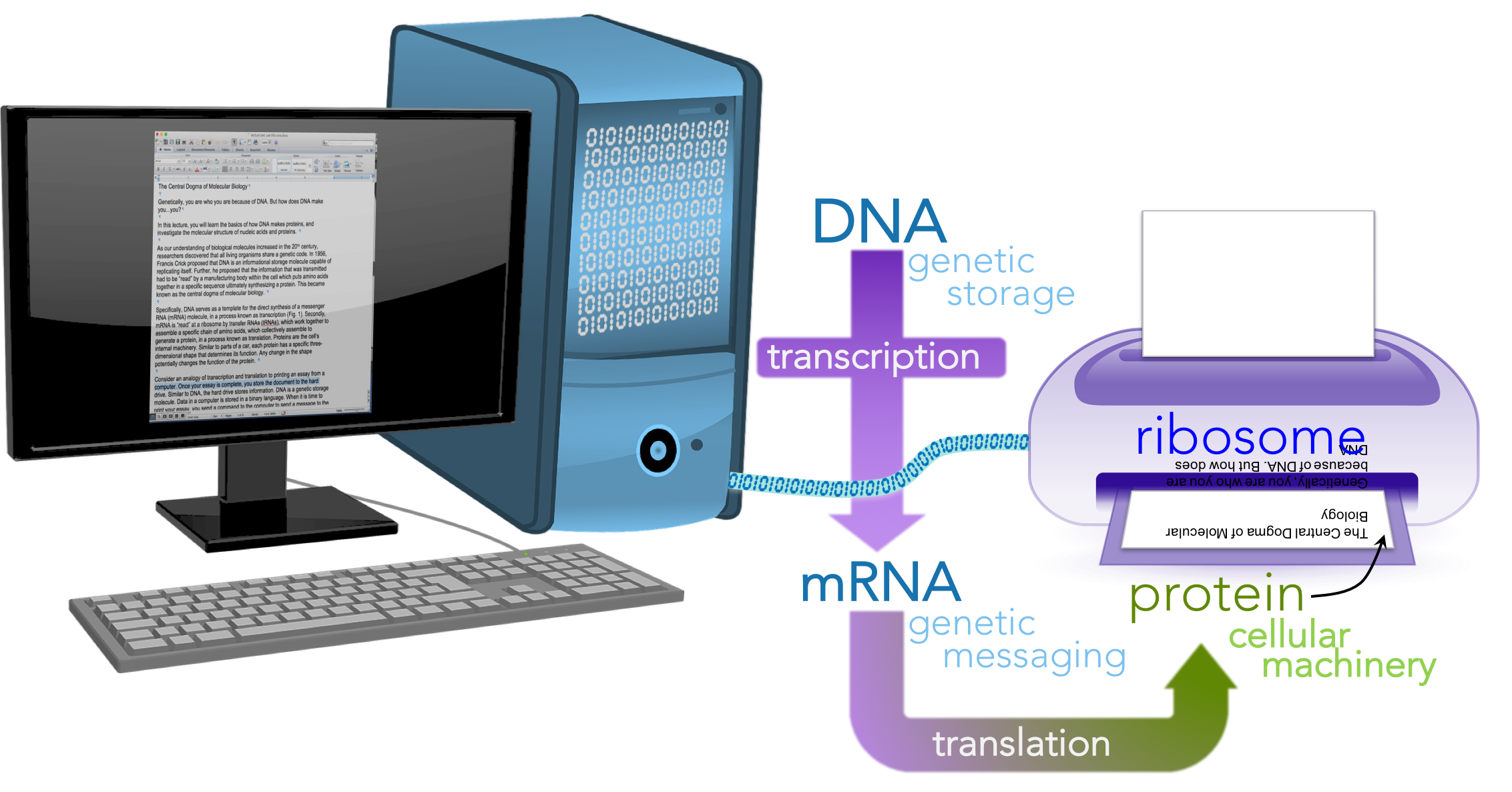
Consider an analogy of transcription and translation to printing an essay from a computer (Fig. 2).
Figure 2. Printing a paper as a metaphor for the central dogma.
Once your essay is complete, you store the document to the hard drive. Similar to DNA, the hard drive stores information. DNA is a genetic storage molecule. Data in a computer is stored in a binary language. When it is time to print your essay, you send a command to the computer to send a message to the printer. This message is akin to mRNA, a genetic messaging molecule. Similar to the binary message sent to the printer, DNA and RNA share a chemical language (based on nucleotides), hence why the information exchange from DNA to RNA is called transcription, an exchange of information in the same language. Once the information is received at the printer, it is translated from binary language into a different language, a language of ink. Analogously, mRNA is read by the ribosome and translated into the language of proteins, which are made up of amino acids. Thus, the process from RNA to proteins is known as translation, translating from the language of nucleotides to amino acids. The ribosome is akin to the printer, serving as a facility for the process of translation. The molecules that actually translate the mRNA at the ribosome are a different kind RNA, transfer RNA or tRNA. In the process of translation, a tRNA reads the mRNA and links a specific amino acid to a growing protein. For your essay to represent your idea, the ink must be physically arranged in a specific manner. Any malfunction in the positioning of letters would not convey the same idea. Similarly, proteins have a specific three-dimensional shape that determines their function. Any change in that shape can potentially alter its function. In our analogy DNA is the stored file in the hard drive; mRNA is message sent to the printer; the printer is the ribosome; the essay is the protein and the letters represent amino acids.
DNA indirectly codes for proteins. DNA directly creates all of the intermediate players of transcription and translation. DNA’s day-to-day function is the production of RNA molecules. Messenger RNA (mRNA) is directly generated by a specific segment of DNA. That segment of DNA is known as a gene. The mRNA travels to a ribosome, which is made up of protein and another type of RNA, ribosomal RNA (rRNA). At the ribosome, the mRNA serves as a code for the synthesis of protein by linking specific amino acids in an exact sequence. The overall collection of an amino acid chain is a protein. DNA is also capable of self-replication, necessary for the creation of new cells.
Nucleic Acids
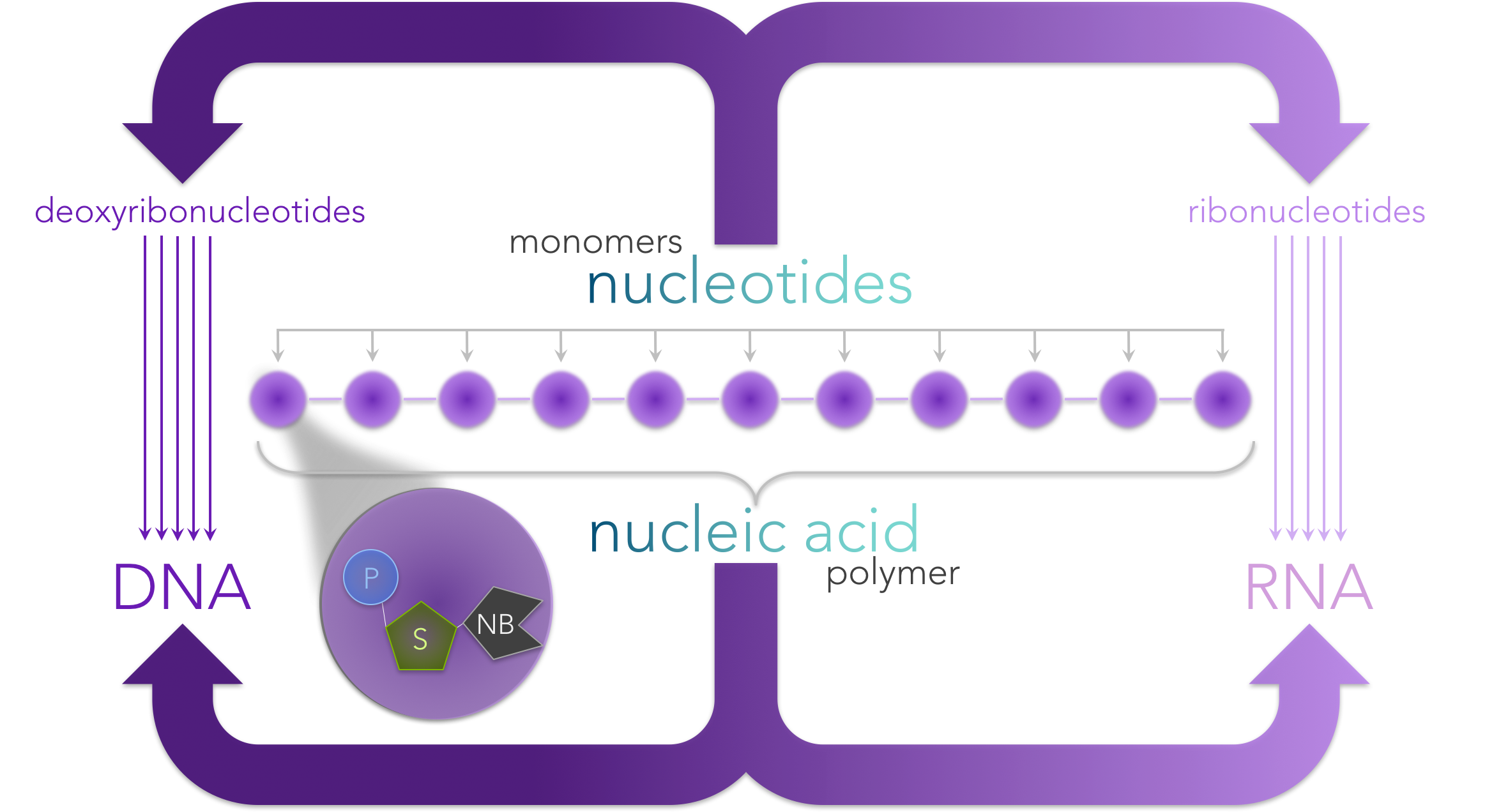
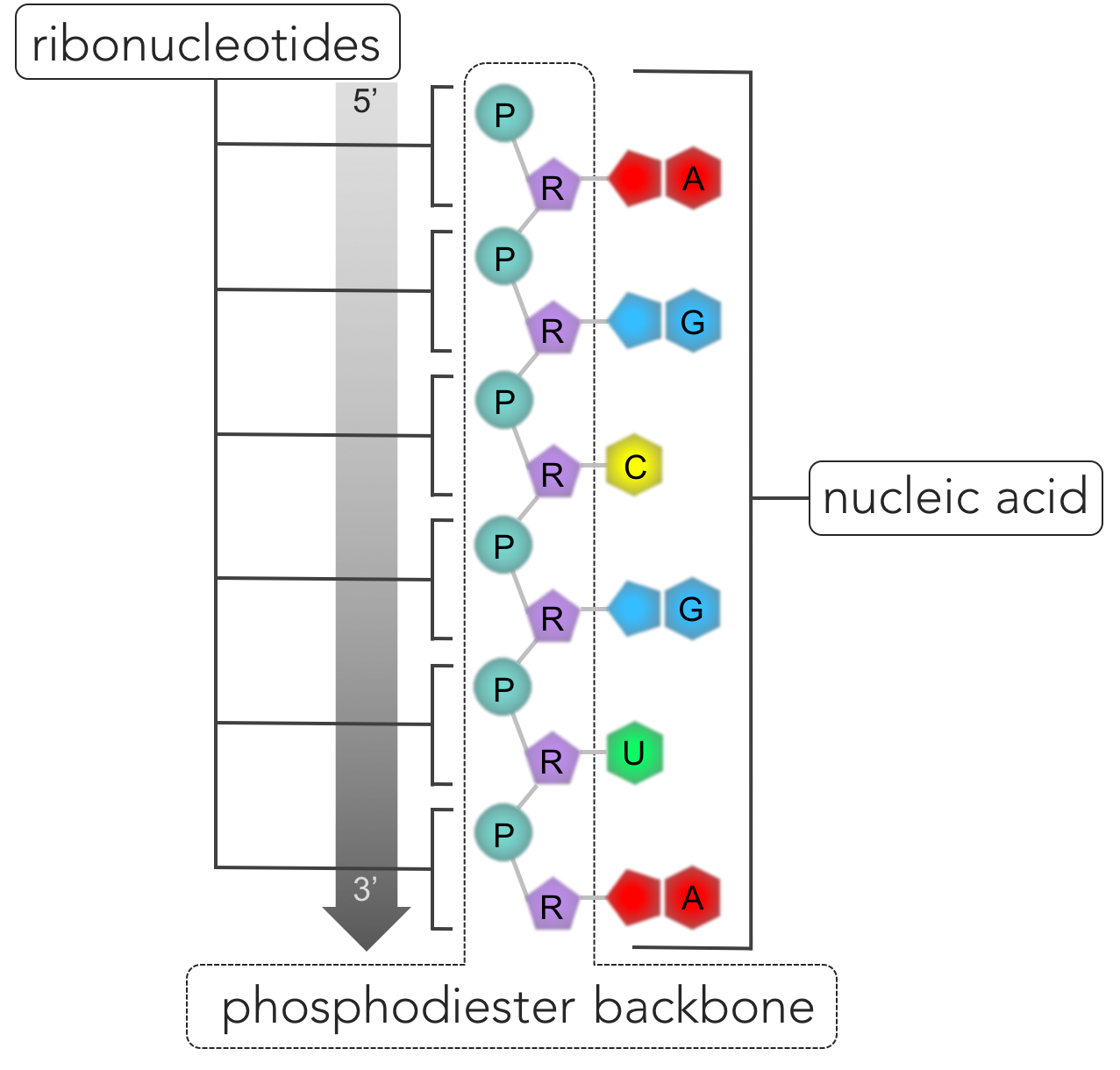
Figure 3. Nucleic acids are polymers made of a chain of nucleotide monomers. Each nucleotide is composed of a phosphate (P), sugar (S) and a nitrogenous base (NB). Ribonucleotides have the sugar ribose, whereas deoxyribonucleotides have deoxyribose. The nitrogenous bases for ribonucleotides are adenine (A), uracil (U), cytosine (C) and guanine (G), whereas deoxyribonucleotides have the nitrogenous based A, C, G and thymine (T).
DNA and RNA are biological molecules known as nucleic acids (Fig. 3). Nucleic acids (as well as proteins) are polymers, or molecules made up of a linking chain of repeating molecules. The repeating components are known as monomers. The monomers of nucleic acids are nucleotides, which are composed of three components: a sugar, a phosphate group and a nitrogenous base.
Deoxyribonucleotides
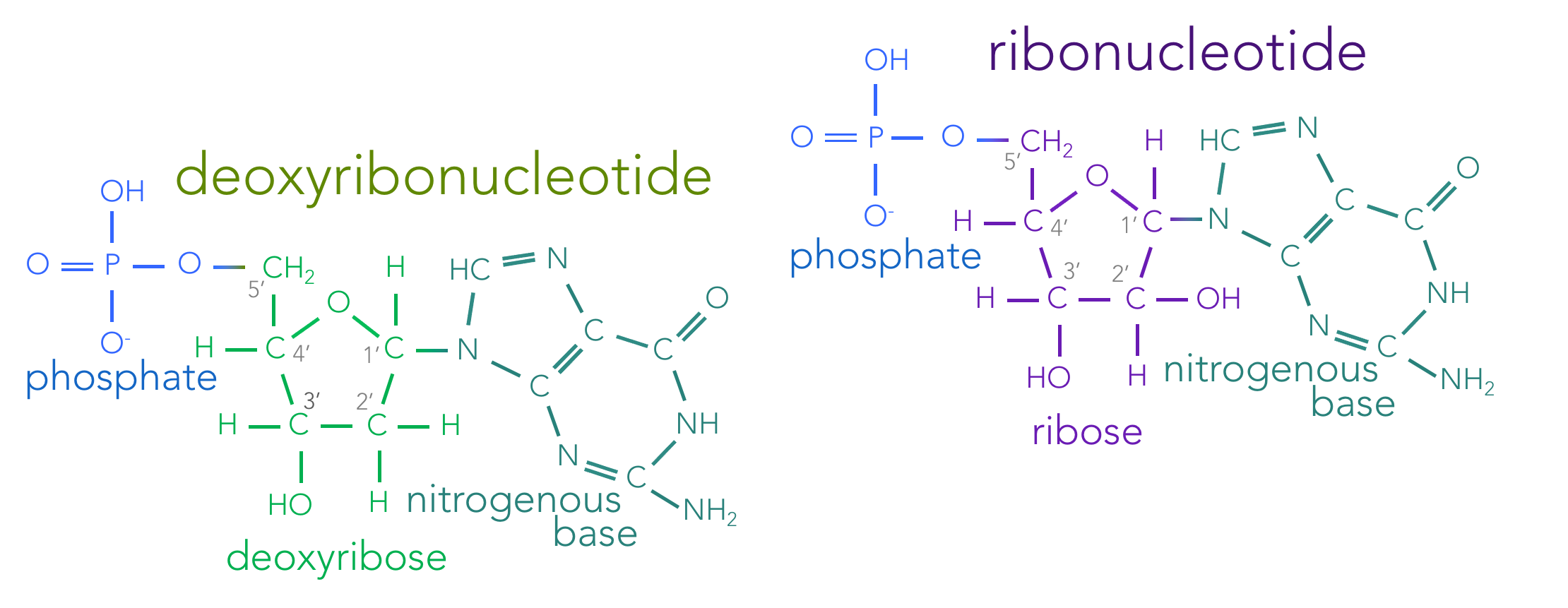
DNA (or deoxyribonucleic acid) is a double-stranded nucleic acid composed of monomers known as deoxyribonucleotides. A deoxyribonucleotide is made up of three components: a phosphate group, the sugar deoxyribose, and one of four nitrogenous bases (Fig. 3).
While the phosphate group and deoxyribose are identical in the varying deoxyribonucleotides, DNA houses four different nitrogenous bases: adenine (A), thymine (T), cytosine (C), and guanine (G). These four different nucleotides serve as the letters of the genetic informational storage, which are transcribed into mRNA and eventually read at the ribosome to create a protein. All the biological diversity on earth in the world is based on the language of life, which only has four letters. Nitrogenous bases can be placed into to categories, based on their shape. Thymine and cytosine are each composed of a single carbon ring skeleton, and are known as pyrimidines; whereas, adenine and guanine are composed of two carbon ring skeletons connected together (one six-sided and the other five-sided), and these are known as purines.
Ribonucleotides
Figure 4. Skeletal structure of nucleotides. Nucleotides are composed of three molecular components: a phosphate, a sugar and a nitrogenous base. Living organisms contain both deoxyribonucleotides (which combine to form DNA) and ribonucleotides (forming RNA). Deoxyribonucleotides contain the sugar deoxyribose, where as ribonucleotides contain the sugar ribose. These sugars are identical, except the presence of an oxygen on the 2' carbon in ribose. Deoxyribonucleotides have one of four nitrogenous bases: adenine, thymine, cytosine or guanine. Ribonucleotides have the nitrogenous bases: adenine, uracil, cytosine and guanine.
Ribonucleic acids (RNAs) are single stranded nucleic acid polymers made up of the monomers, ribonucleotides (Fig. 4). Ribonucleotides are nearly identical to deoxyribonucleotides with two exceptions.
First, ribonucleotides are made of the sugar ribose, which has a hydroxide (OH) at the 2’ carbon, whereas deoxyribose a hydrogen (H) atom at that location. The carbons of ribose and deoxyribose are notated moving clockwise from the oxygen in the ring: 1’ to 5’. The 1’ carbon connects to the nitrogenous base. The atoms attached to the 2’ carbon differ between ribose and deoxyribose. Clockwise from the 2’ carbon is the 3’ carbon, followed by the 4’ carbon. And the 5’ carbon attaches to the phosphate group. So relative to RNA’s sugar, ribose, DNA’s sugar, deoxyribose, lacks an oxygen on the 2’ carbon, hence deoxyribose. That single difference allows cells to differentiate between the two nucleotides.
Second, ribonucleotides differ in their suite of nitrogenous bases. Three ribonucleotides have the same nitrogenous bases as deoxyribonucleotides: cytosine, guanine and adenine. While the fourth ribonucleotide is composed of the nitrogenous base: uracil (U). Uracil is very similar to thymine, except there is a hydrogen atom at the 3’ location of uracil, while thymine has a methyl (CH3) group there. The phosphate group is identical for both ribonucleotides and deoxyribonucleotides.
Phosphodiester Bond
Figure 5. Phosphodiester bond. The phosphodiester bond between two nucleotides is a covalent bond between the 3' carbon of one sugar and the phosphate of an additional sugar.
Nucleotides link together in long chains to form a nucleic acid. Individual nucleotides are connected by a covalent bond that forms between the 3’ carbon (C) of the sugar molecule of one nucleotide and a phosphorous (P) of the phosphate group of an adjacent nucleotide (Fig. 5). In this reaction, a hydrogen atom is removed from the 3’ carbon and hydroxyl (OH) is removed from the phosphate. These byproducts combine forming water (H2O), in a reaction known as a condensation reaction. Following this reaction the two nucleotides are connected by a phosphodiester bond, in which a phosphate group (PO4) is linked to the 5’ carbon of its original nucleotide and the 3’ carbon of an adjacent nucleotide.
Nucleic backbone
Figure 6. RNA. Nucleic acids are polymers composed of a single strand (as in RNA) or two strands (DNA) of nucleotides connected by phosphodiester bonds. The repeating pattern of connected phosphate groups (P) and sugars (R) connected form the phosphodiester backbone, while the nitrogenous bases (A, U, C, and G in the case of RNA) hang off the side. One side of the strand of the nucleic acid is bounded by a phosphate group (denoted the 5' end) and a sugar group is located on the opposite end (denoted 3').
Adding a third nucleotide, the nucleic acid begins to take shape. In this developing nucleic acid, a phosphate is attached to a sugar, which is attached to a phosphate attached to sugar, and so on. A phosphodiester linkage only involves two of the three components of a nucleotide: phosphate and sugar. Hanging off to the side of the nucleic backbone are the nitrogenous bases. This repeating pattern forms the backbone of nucleic acids. One end of a nucleic acid strand is bound by a phosphate group, while the opposite end is bound by a sugar, giving DNA and RNA directionality. The phosphate group terminus of a nucleic acid is referred to as the 5’ end of the strand, as the 5’ carbon is the closest carbon to the end of the molecule. The opposite end of the nucleic backbone contains a sugar terminus, called the 3’ end. The sequence of nucleotides in a nucleic acid is known as its primary structure. Scientists have standardized the notation of nucleic acids primary structure by listing the nucleotides from the 5’ end to the 3’ end (5’to 3’). For example, a segment of RNA (adenine-guanine-guanine-uracil-adenine-cytosine), would be notated: AGGUAC.
RNA
RNA (ribonucleic acid) is a single-stranded nucleic acid, composed of ribonucleotides (Fig 6). The nucleic backbone of RNA is bound by a phosphate group on the 5’ terminus and ribose (a sugar) on the 3’ terminus. Each ribonucleotide has one of four nitrogenous bases: adenine, uracil, cytosine and guanine. Ribonucleotides combine via phosphodiester bonds to form RNA.
RNA molecules are predominately responsible actively synthesizing proteins. DNA synthesizes messenger RNA (mRNA), which transmits genetic information from DNA to a ribosome. The primary sequence of the mRNA determines the sequence of amino acids in the resultant protein. Ribosomes are hybrid complexes made of proteins and a different type of RNA, ribosomal RNA (rRNA). At the ribosome, the mRNA is read and decoded by a third RNA, transfer RNA (tRNA). In eukaryotes, small nuclear RNA (snRNA) is involved in modifying mRNA after transcription and before translation.'
DNA
DNA (deoxyribonucleic acid) is a double-stranded nucleic acid composed of deoxyribonucleotides (Fig. 4), which vary from ribonucleotides by having a different 5-carbon sugar, called deoxyribose. In living organisms, there are four deoxyribonucleotides that vary in their nitrogenous bases: adenine, thymine, cytosine and guanine.
Figure 7. Chargaff's discovery. Erwin Chargaff discovered that the abundance of of cytosine and guanine are equal to each other & the abundance of adenine and thymine are equal within an organism. He found these results consistent across several species, and lead James Watson and Francis Crick to hypothesize DNA base pairing hypothesis.
Once the primary structure of DNA was solidified, the next question was how are the nucleotides arranged to create the DNA molecule, or the secondary structure of DNA. The discovery of DNA’s secondary structure was one of the most important biological discoveries in the 20th century. One of the first clues came from analyses conducted in the early 1950s by Erwin Chargaff comparing the relative abundances of deoxyribonucleotides across a variety of organisms (Fig 7). Chargaff discovered: 1) the relative abundance of guanine equals cytosine, and 2) the relative abundance of adenine equals thymine. And what is most interesting about this is that he found this relationship across many different species of organisms. Chargaff’s discovery was instrumental to scientists uncovering DNA’s secondary structure. James Watson and Francis Crick suggested Chargaff’s evidence strongly supports base pairing in DNA, in which deoxyribonucleotides of adenine attach to thymine (A↔T) and guanine attaches to cytosine (G↔C). In addition, Watson and Crick hypothesized that base pairing of deoxyribonucleotides suggested that DNA was most likely double stranded.
Figure 8. X-ray crystallography revealed DNA's molecular shape. Rosiland Franklin, working with Maurice Wilkins developed techniques to produce an image that revealed that DNA has a consistent width, a repeating pattern of nucleotides and is helical.
To acquire evidence of the actual molecular shape of DNA, Rosalind Franklin and Maurice Wilkins bombarded DNA with x-rays and analyzed how the radiation scattered, a technique known as x-ray cyrstallography (Fig. 8). Analyses of the scatterplots from this technique allowed them to measure the distance between atoms in DNA and they were able to conclude three things: 1) DNA has a consistent width, 2) within DNA is a repeating pattern and 3) the molecule must be helical. In collaboration with Franklin and Wilkins, Watson and Crick used the measurements to define the geometry of the components of deoxyribonucleotides. Creating physical models of the nucleotides (literally paper cut-outs), Watson and Crick tinkered with different arrangements of the nucleotides to explain the 1) Chargaff’s rule, 2) a consistent width, 3) the repeating pattern of the nucleotides and 4) helical shape of DNA.
Figure 9. Secondary structure of DNA. James Watson and Francis Crick uncovered the secondary structure of DNA. 1) DNA is double stranded. 2) The strands are antiparallel. 3) The strands are held together by base pairing of hydrogen bonds between the nitrogenous bases, where adenine binds with thymine and cytosine binds with guanine.
By arranging base pairing nucleotides (A↔T & G↔C) side by side in strands running in opposite directions, all of the discoveries could be explained (Fig 9). Watson and Crick suggested that DNA is composed of two strands: one running 5’ to 3’ connected to a second strand running 3’ to 5’. This orientation is called antiparallel. The nucleic backbone is composed of alternating phosphate and deoxyribose sugar molecules with a phosphate on the 5’ end of the strand and a deoxyribose on the 3’ end. The strands twist to form a double helix, a spiral bounded on the outside by two nucleic backbones running in opposite directions, with the nitrogenous bases facing inward.
Based on Chargaff’s findings, Watson and Crick determined that the nitrogenous bases from adjacent DNA strands connect according on base pairing. The discovery of a consistent width of the DNA molecule also supported the A-T and C-G base pairing. While adenine and guanine are different molecules, they are both purines and approximately the same size and shape. The same is true of the pyrimidines, cytosine and thymine. However, purines consist of a figure eight structure, which is larger than the circular structure of pyrimidines. For the width of DNA to be consistent with the variety of shapes found in nitrogenous bases, purines must connect with pyrimidines. A purine-purine base pairing creates a larger molecular width than observed, and a pyrimidine-pyrimidine base pairing would be too small.
Figure 10. Base pairing in DNA. Deoxyribonucleotides connect to adjacent deoxyribonucleotides based on complementary base pairing. DNA strands connect via hydrogen bonds. Adenine and thymine form two hydrogen bonds. Guanine and cytosine bind with three hydrogen bonds.
If purine-pyrimidine base pairing explains the consistent width of a double-stranded DNA molecule, why does guanine (a purine) appear to always bind with cytosine (a pyrimidine) but never thymine (also a pyrimidine)? Why doesn’t adenine (a purine) bind with cytosine? With their physical models of nucleotides, Watson and Crick deduced the nitrogenous bases of adjacent strands were held together by hydrogen bonding (FIg. 10). Due to the differential in electronegativities, the hydrogens of the nitrogenous bases are partially positive (δ+), and the oxygens and nitrogens are partially negative (δ-). Hydrogen bonds form between the δ+ and δ- atoms of adjacent nitrogenous bases. Investigating the shapes and interactions of these four nitrogenous bases, they discovered that guanine and cytosine were geometrically complements of each other and held together by three hydrogen bonds, while adenine and thymine are held together by two hydrogen bonds. Essentially, the A-T and C-G pairing are more stable than any other combination due to the complementarity of the molecular shape and hydrogen bond orientation.
Proteins
Proteins are biological molecules that serve as cellular machines in living organisms. These large molecules are specific three-dimensional structures involved in biological processes such as cellular signaling, catalyzing chemical reactions, molecular transportation, and many other functions. Proteins are polymers, consisting of long chains of monomers, amino acids.
Amino acids
Figure 11. Primary structure of an amino acid. An amino acid is composed of an amine group, a central carbon, a carboxyl group and an R-group. R-groups vary from amino acid to amino acid.
Of the more than 500 amino acids known, only 20 appear in proteins of living organisms. An amino acid is a relatively simple organic molecule (Fig 11). Attached to a central carbon by single covalent bonds are: 1) a hydrogen atom, 2) an amine group (NH3+), 3) a carboxylic acid (COO-) group and 4) an R-group, also known as a side chain.
At around pH 7 as in water, the amine group of an amino acid attracts a proton becoming NH3+, and acts as a base. The carboxyl group is negatively charged in water, due to the high electronegativity of both oxygens pulling electrons from hydrogen and losing the proton. Different amino acids vary in their R-group. Of the protein-building amino acids, the R-groups can vary in their size, shape and polarity. Proteins, being made up of chains of amino acids, vary based on the interactions of the atoms within the amino acids and water. These interactions dictate the shape of the protein, which in turn determines its function.
R-groups vary in their polarity. Non-polar molecules have relatively equal distribution of electrons via covalent bonding, while polar molecules have an unequal distribution of electrons. The unequal distribution of electrons in polar molecules creates partially charged atoms (δ+, δ-). Polar R-groups are hydrophilic, meaning they have an affinity for water due to hydrogen bonds between the partial charges of the R-group and the water molecules. Non-polar R-groups are repelled by water, or hydrophobic. Therefore in a chain of amino acids, ones with polar R-groups will bend towards water and non-polar R-groups bend away, affecting the eventual shape of a protein.
Peptide bonding
Figure 12. A polypeptide is composed of several amino acids connected by peptide bonds. Note: on one end of the polypeptide is an amine group, whereas a carboxyl group is on the opposite end.
Proteins are polymers of amino acids chained together by covalent bonds, known as peptide bonds (Fig 12). A peptide bond is a condensation reaction, in which the oxygen ion (O-) of the carboxylic acid from one amino acid is removed (becoming carboxyl) and combines with two hydrogen atoms (2H) from the amine group of an adjacent amino acid to produce water (H2O). A covalent bond forms between two amino acids when the carbon of the carboxyl group that lost the OH during the condensation reaction combines with the adjacent nitrogen (N) of another amino acid that lost the hydrogen atoms, bonding two adjacent amino acids. This is a peptide bond. Amino acids link via peptide bonds forming long chained molecules, or polypeptides.
Levels of protein structure
Proteins have four levels of structure.
Primary structure
Figure 13. Sickle cell anemia is caused by an alteration of hemoglobin's primary structure. An alteration in a single deoxyribonucleotide alters the transcribed mRNA. This alters one amino acid in the protein, hemoglobin (responsible for binding to oxygen), creating a sickle-shaped red blood cell.
The three dimensional shape of a protein determines its function, and the shape of proteins are ultimately dependent upon the sequence of amino acids coded for by DNA. The unique amino acid sequence is a protein’s primary structure.
Figure 14. Geographical distribution of sickle cell anemia and malaria.
In humans, sickled cell anemia is an inherited condition in which blood cells have a variant of the oxygen-binding protein, hemoglobin. Sickle-celled anemia is considered a disease of the primary structure of proteins as it is caused by the variation of a single amino acid in hemoglobin: a valine instead of a glutamate in the sixth position of a 146 amino acid protein (Fig. 13). While normal blood cells are rounded, humans with this variant produce sickle-shaped red blood cells. Normal blood cells are elastic and flow freely through veins, but sickled red blood cells are rigid and tend to get stuck where veins branch. This blockage starves downstream tissues of oxygen resulting in a host of medical issues, including lower life expectancy. The body identifies the cells when they get stuck and destroys them. Healthy red blood cells typically live between 90-120 days, whereas a sickled red blood cells have a 10-20 day life span. Therefore, people with sickle-celled anemia must produce much more blood, which is rich in iron, leading to an overall iron deficiency, or anemia. Interestingly, sickle-cell anemia is evolutionarily advantageous in certain circumstances. Prior to globalization, the highest rates of sickle-celled anemia occur in tropical Africa, Middle East and India, all malaria-dominated areas (Fig. 14). Malaria is a single-celled eukaryotic parasite transmitted by mosquitoes that some consider the most deadly human disease ever. However, in sickled red blood cells the malaria parasites cause the cell to rupture before they can successfully reproduce. Therefore, people with sickle cell anemia have an evolutionary advantage or people with normal blood cells in these areas. In areas absent of malaria, sickled red blood cells are highly disadvantageous due to the host of medical conditions associated with this variant. All of this is caused by a single different amino acid in one protein, an alteration of the proteins primary structure.
Secondary structure
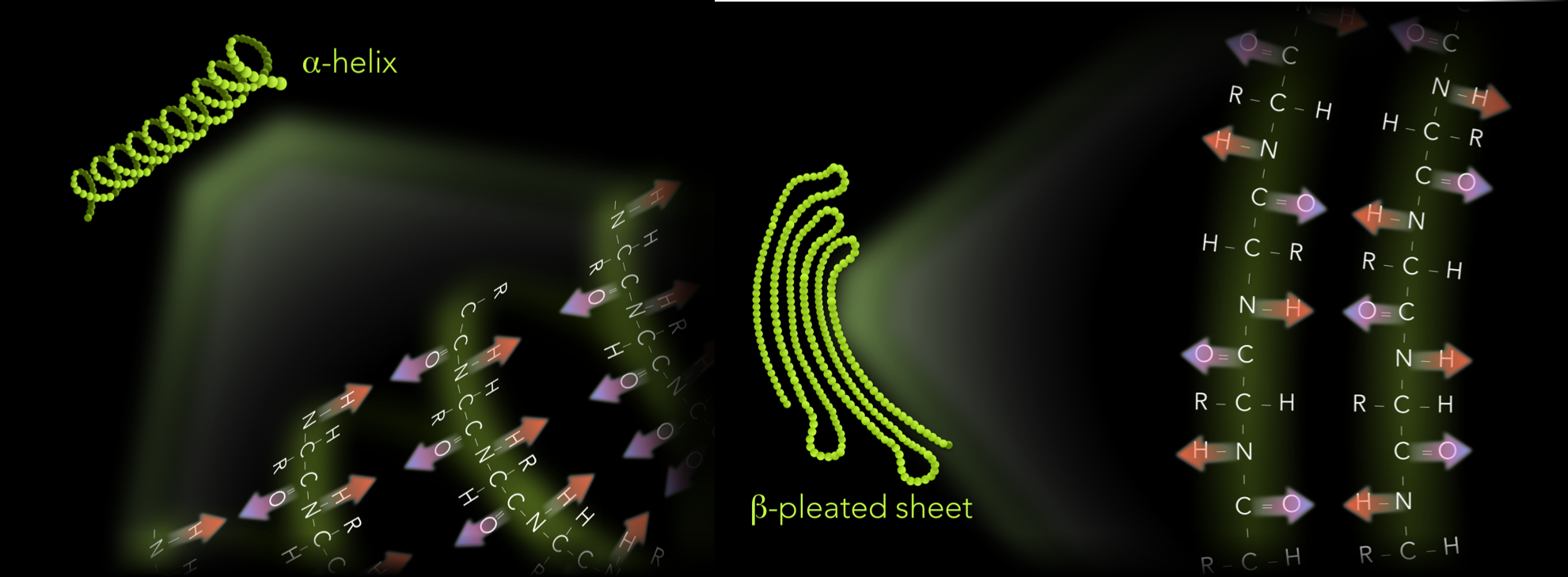
Figure 15. Primary and secondary structure of proteins. A protein's primary structure is the sequence of amino acids. Secondary structure is created by repeating molecular interactions between the carboxyl and amino groups, forming either alpha-helices or beta-pleated sheets.
When amino acids are grouped into polypeptide chains, neighboring amino acids can interact via hydrogen bonding. These interactions can form a regular pattern, which increases the molecular stability of the polypeptide chain. These patterns are known as the protein’s secondary structure and form either corkscrew-shaped structures, known as α-helices, or folded ribbon-shaped structures, known as β-pleated sheets (Fig 15).
Recall oxygen has high electronegativity, while hydrogen has low electronegativity. This differential in electronegativity results in hydrogen bonding between neighboring amino acids. Within a polypeptide backbone, hydrogen bonds can occur between amine groups and carboxyls. The partially negative oxygen of the carboxyl can bind with the hydrogen of amine groups of other amino acids.
In α-helices and β-pleated sheets, hydrogen bonding occurs between amine and carboxyl groups of different amino acids (Fig 16). How the hydrogen bonding occurs between amino acids determines which of the two shapes emerges. Within a single amino acid of an α-helix polypeptide chain, the hydrogens of the amine groups face the opposite direction relative to the oxygens of the carboxyl groups. Down the entire α-helix, the hydrogens of the amine group face the same direction, and oxygens of the carboxyl group orient in the opposite direction. Within the α-helix, a hydrogen bond forms when an oxygen of a carboxyl faces a hydrogen of a different amino acid further down the polypeptide chain. This accumulation of multiple hydrogen bonds stabilizes the polypeptide chain.
β-pleated sheets are also formed by hydrogen bonding between the amine and carboxyl groups of different amino acids. However, the orientation of these groups differs. In a single amino acid of a β-pleated sheet, the oxygen of the carboxyl and the hydrogen of the amine group face in the same direction. In the adjacent amino acid, the oxygen and hydrogen both face in the opposite direction relative to the first. The third amino acid is in a similar orientation to the first, and so on. In β-pleated sheets, hydrogen bonds also occur between the carboxyl oxygen and amine hydrogen between neighboring amino acids. However, the orientation of these atoms causes the structure to bend into a folded ribbon shape, or β-pleated sheet.
Figure 16. Hydrogen bonding between amino and carboxyl groups determine a protein's secondary structure. Alpha-helices and beta-pleated sheets emerge as hydrogen bonding occurs between the partially negative oxygens of the carboxyl groups and partially negative hydrogens of the amine groups. In alpha-helices, the carboxyl oxygen faces the opposite direction relative to the amine hydrogen within a single amino acid. Along the polypeptide, all the amine hydrogens face the same direction. Carboxyl oxygens all face the opposite direction. In beta-pleated sheets, the carboxyl oxygen and amine hydrogen within a single amino acid, face the same directions. Oxygens and hydrogens of adjacent amino acids face in opposite directions.
Tertiary structure
While the secondary structure of proteins is determined by the interactions between amine groups and carboxyl groups of neighboring acids, tertiary structure is defined by how the R-groups of neighboring amino acids interact (Fig. 17). These interactions result in very specific folding patterns eventually helping to stabilize a specific 3-dimensional structure of the polypeptide. Several types of interactions occur between neighboring R-groups.
Figure 17. Tertiary structure of proteins. a) Tertiary structure is affected by how R-groups interact with each other and water. b) Some R-groups are either polar or non-polar. Partially negative atoms (O & N) of polar R-groups form hydrogen bonds with partially positive hydrogens of neighboring R-groups. c) Polar R-groups are hydrophillic, bending towards water. Whereas, non-polar R-groups are hydrophobic and bend away from water.
Hydrogen bonding
While the hydrogen bonding determines the secondary structure of proteins, hydrogen bonding can also occur between the R-groups of a polypeptide chain. The 20 R-groups of amino acids are either polar or non-polar. Polar R-groups have oxygen or nitrogen atoms, which characteristically have high electronegativity due to their high affinity for electrons. These polar R-groups tend to bond with hydrogen atoms of neighboring non-polar R-groups or the hydrogen of the amine group of the peptide backbone. Non-polar R-groups can also form hydrogen bonds with the peptide backbone, either by interacting with the oxygen of the carboxyl group or the nitrogen of the amine group. While hydrogen bonding is relatively weak, the overarching abundance of these interactions forms very stable polypeptide structures.
Hydrophobic and hydrophilic interactions
Figure 18. Quaternary structure of a protein.
In living organisms, proteins are surrounded by water. Polar R-groups are hydrophilic and bend to turn towards the water, whereas non-polar R-groups are hydrophobic and turn away from water. Hydrophobic R-groups tend to amass in the internal section of the protein forming globular masses.
Ionic bonding
While hydrogen bonding is facilitated by the interactions of partial charges, certain R-groups have full charges and are involved in ionic bonding. This happens between completely positive R-groups form ionic bonds with neighboring R-groups that are completely negative.
Quaternary structure
The overall structure of a fully-functional protein is known as the quaternary structure (Fig. 18). Most proteins are composed of several polypeptides. A polypeptide is composed of either a series of α-helices with tertiary level interactions, or a series of β-pleated sheets with tertiary level interactions.
© 2016 Jason S. Walker. All rights reserved.